Translate this page into:
Hereditary hypotrichosis simplex with SNRPE gene mutation
Corresponding author: Dr. Hyun-Chang Ko, Department of Dermatology, School of Medicine, Pusan National University, Seo-Gu, Busan, Republic of Korea, Korea, hcko@pusan.ac.kr
-
Received: ,
Accepted: ,
How to cite this article: Won Y, Kim B, Kim M-B, Ko H-C. Hereditary hypotrichosis simplex with SNRPE gene mutation. Indian J Dermatol Venereol Leprol. doi: 10.25259/IJDVL_191_2024
Dear Editor,
Hereditary hypotrichosis simplex (HHS) is a rare form of non-syndromic hereditary hypotrichosis. Individuals exhibit normal hair at birth but experience progressive loss and thinning in early childhood, continuing as they age. Scalp and body hair involvement varies widely among affected individuals and within families.
HHS results from various genes with autosomal dominant or recessive inheritance. Due to the genetic heterogeneity observed in non-syndromic hypotrichosis, phenotypic classifications have been established, known as hypotrichosis 1 through to hypotrichosis 15, based on associated genes and gene loci. This system enhances the understanding of diverse manifestations of different genetic factors. Hypotrichosis 11 stems from a heterozygous SNRPE gene mutation on chromosome 1q32.
A 27-month-old boy presented with progressive hair thinning since birth [Figure 1a]. He had black hair at birth, which gradually thinned and turned brown. There were no eyebrows or eyelashes [Figure 1b]. His nails, teeth, and skeletal radiographs were normal, with no developmental delays. The boy’s Korean father had a similar clinical history, experiencing hair thinning after one year of age. During the visit, the father exhibited slight, light brown hair on the temporal scalp but no hair over eyebrows, eyelashes, body, or pubic region [Figures 2a and 2b]. No other reported hair loss symptoms were evident in paternal or maternal family members. The Exome sequencing unveiled a heterozygous c.1A>G (p.Met1?) mutation, confirmed by Sanger sequencing [Figure 3]. This variation is anticipated to be pathogenic as predicted by the computational tool MutationTaster (http://www.mutationtaster.org/), potentially disrupting the initiation codon of the SNRPE gene and triggering subsequent activation of alternative translation initiation sites, leading to a shortened amino acid sequence.
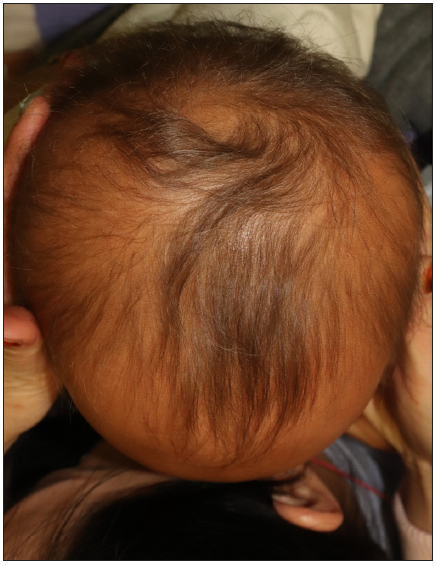
- Top view of the patient’s scalp.
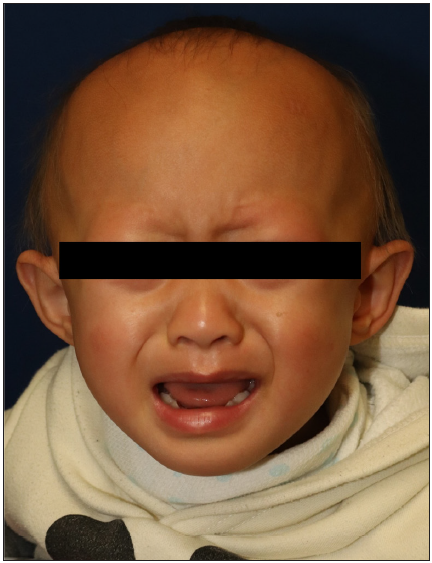
- Front view of the patient showing sparse, thin hair, with absence of eyebrows and eyelashes.
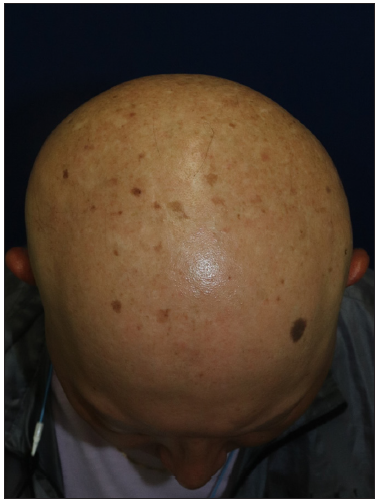
- Top view of the father’s scalp.
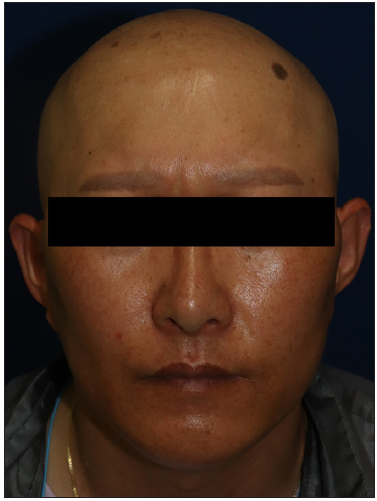
- Front view of the father with tattooed eyebrows.
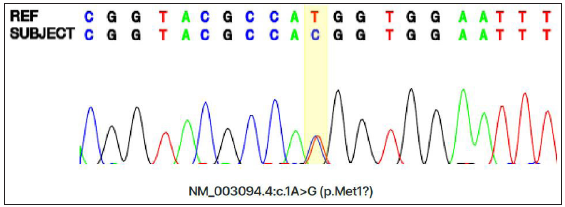
- Sequence analysis of SNRPE in the patient revealed a c.1A>G substitution. (A: Adenosine, C: Cytosine, T: Thymine, G: Guanine).
The SNRPE protein is crucial for pre-mRNA (messenger ribonucleic acid) processing, especially in assembling uridine (U)-rich small nuclear ribonuclear proteins (UsnRNPs). Heterozygous SNRPE gene variants have been found in eight families. Pasternack et al.1 reported Spanish and the UK families with the c.1A>G (p.Met1?) mutation and one Turkish family with the c.133G>A (p.Gly45Ser) mutation, all displaying isolated hypotrichosis simplex. Additionally, the same research group reported another Turkish family with the c.1A>G (p.Met1?) mutation, along with two other families exhibiting the c.54+2T>A and c.221T>C (p.Leu74Pro) mutations – a Chinese family and a Spanish family, respectively, all associated with the isolated hypotrichosis simplex.2 In contrast, Chen et al.3 reported that the c.65T>C (p.Phe22Ser) mutation was linked to isolated non-syndromic congenital microcephaly and intellectual disability. Hypotrichosis was not mentioned or documented in this patient. Amudhavalli et al.4 documented a c.82-28_82-16del (p.Arg28_Ile48del) mutation leading to microcephaly, congenital atrichia and multiple anomalies. Among them, the c.1A>G mutation was detected in only three families from Spain, the United Kingdom, and Turkey, making ours the fourth instance.
Pan et al. noted a limited number of reported SNRPE mutations across diverse ethnicities2, which is intriguing. A Spanish family with six members carrying the c.1A>G mutation exhibited a highly variable degree of alopecia since birth.1 We propose that variable expressivity may pose challenges for physicians in recognising symptoms, potentially contributing to the small number of reported cases despite the mutation being observed in four diverse ethnicities. Our research contributes to expanding the ethnic variability of SNRPE gene mutations, particularly highlighting the occurrence of the c.1A>G mutation in the Asian population, implying this locus is a potential hotspot. The accumulation of such data is significant, as it helps document common disease-causing hotspots across various ethnic groups.
In light of our findings, dermatologists should prioritise the consideration of HHS due to its genetic diversity, particularly in cases where conventional treatments are ineffective and there is a family history of progressive hair thinning.
Declaration of patient consent
The authors certify that they have obtained all appropriate patient consent.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Use of artificial intelligence (AI)-assisted technology for manuscript preparation
The authors confirm that there was no use of AI-assisted technology for assisting in the writing or editing of the manuscript and no images were manipulated using AI.
References
- Mutations in SNRPE, which encodes a core protein of the spliceosome, cause autosomal-dominant hypotrichosis simplex. Am J Hum Genet. 2013;92:81-7.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Additional causal SNRPE mutations in hereditary hypotrichosis simplex. Br J Dermatol. 2021;185:439-41.
- [CrossRef] [PubMed] [Google Scholar]
- A missense mutation in SNRPE linked to non-syndromal microcephaly interferes with U snRNP assembly and pre-mRNA splicing. PLoS Genet. 2019;15:e1008460.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Novel blended SNRPE-related spliceosomopathy phenotype characterized by microcephaly and congenital atrichia. Am J Med Genet A. 2023;191:1425-9.
- [CrossRef] [PubMed] [Google Scholar]





