Translate this page into:
Resistance to anti leprosy drugs in multi-bacillary leprosy: A cross sectional study from a tertiary care centre in eastern Uttar Pradesh, India
2 Department of Microbiology, Institute of Medical Sciences, Banaras Hindu University, Varanasi, Uttar Pradesh, India
3 Division of Biostatistics, Institute of Medical Sciences, Banaras Hindu University, Varanasi, Uttar Pradesh, India
Correspondence Address:
Satyendra Kumar Singh
Department of Dermatology and Venereology, Institute of Medical Sciences, Banaras Hindu University, Varanasi, Uttar Pradesh
India
| How to cite this article: Singh SK, Kumar A, Nath G, Singh TB, Mishra MN. Resistance to anti leprosy drugs in multi-bacillary leprosy: A cross sectional study from a tertiary care centre in eastern Uttar Pradesh, India. Indian J Dermatol Venereol Leprol 2018;84:275-279 |
Abstract
Background: WHO MDT is the main drug regimen for treating leprosy and has been used for more than three decades. Many cases of relapse of leprosy have been reported, which points towards the emergence of drug resistance with the antileprotic drugs.
Objectives: To find the resistance with the antileprotic drugs by detecting the mutations in drug resistance determining region of the rpoB, folP1 and gyrA genes of Mycobacterium leprae.
Methods: Leprosy patients with bacterial index ≥2 were included in the study. The slides were further processed to extract genomic DNA, and polymerase chain reactions were performed to amplify the drug resistance determining region (DRDR) of rpoB, folP1 and gyrA genes. The samples in which genes could be amplified were subjected to DNA sequencing to detect mutations.
Results: Out of 78 samples rpoB gene was amplified in 39 (50%), folP1 in 32 (41%) and gyrA in 45 (57.7%). In 20 (25.6%) samples no gene was amplified. Only 32 samples of rpoB, 25 samples of folP1 and 38 samples of gyrA gene were included in the study, rest were excluded due to sequencing error. No mutation was seen in rpoB gene and in folP1 gene. In gyrA gene samples mutations were seen in 8 (21%) samples, and were present at codon 91 GCA → GTA (Alanine → Valine).
Limitations: Small sample size and less efficient method to detect resistance.
Conclusion: Resistance is not a problem with conventional drugs in MDT. It is more common with quinolones.
Introduction
Leprosy is a chronic infectious disease which mainly affects skin and peripheral nerves, caused by Mycobacterium leprae (M. leprae). More than 81% of the new cases are reported from three countries – India, Brazil, and Indonesia.[1] Although the prevalence of leprosy in India is less than1/10,000 since 2005,[2] it still accounts for 62% of the total new cases reported worldwide.[1] Dapsone was introduced for the treatment of leprosy in 1950s and was used worldwide to treat both multibacillary (MB) and paucibacillary (PB) forms of the disease. Long-term monotherapy with dapsone resulted in the emergence of dapsone-resistant strains of M. leprae leading to treatment failure.[3],[4] Between 1960s and 1970s, rifampicin and clofazimine were added in the treatment of leprosy. Rifampicin is a strong bactericidal drug against M. leprae. However, using it alone could result in the emergence of rifampicin resistant strains of M. leprae.[5] To overcome the threat posed by the worldwide spread of dapsone and rifampicin resistance and to improve the treatment efficacy, World Health Organization (WHO) recommended multidrug therapy (MDT) for leprosy in 1982. However, drug resistance has been reported since 1964 for dapsone,[3] since 1976 for rifampin,[6] and since 1996 for ofloxacin [7]. Comprehensive data regarding the magnitude of drug resistance is crucial to evaluate the efficacy of MDT. As M. leprae cannot be cultured axenically, detection of drug resistance in leprosy is difficult. Shepard developed mouse foot pad assay to determine M. leprae's susceptibility to anti-leprosy drugs in 1962.[8] Since then, it has been the 'gold standard' for drug susceptibility testing. While mouse foot pad assay gives definitive information pertaining to the susceptibility of an M. leprae isolate to anti-leprosy drugs, it is a laborious and expensive procedure and is carried out only in a few reference centres in the world. The availability of genomic sequence of M. leprae[9],[10] and an improved understanding of the genetic basis of drug resistance in mycobacteria have led to the development of molecular methods for the detection of mutations associated with dapsone, rifampicin, and fluoroquinolone resistance.[11],[12] Recent studies have identified point mutations in the folP1 gene, which encodes dihydropteroate synthase (DHPS) in dapsone-resistant M. leprae.[13] Rifampicin resistance is associated with mutations in the rpoB gene that encodes the β subunit of RNA polymerase.[14] Resistance to ofloxacin is known to be associated with mutation in gyrA gene encoding the A subunit of DNA gyrasein M.leprae.[15],[16] No molecular target has been defined for clofazimine. Thus, by performing polymerase chain reaction and DNA sequencing, we can detect mutations in the drug resistance determining regions (DRDR) of folP1, rpoB, and gyrA genes, responsible for resistance to dapsone, rifampicin and ofloxacin, respectively.[17] The aim of the present study was to look for the reported mutations in folP1, rpoB, and gyrA genes.
Methods
Study design
This was a cross-sectional observational study conducted at a tertiary health care centre during October 2013 to May 2015. The patients included in the present study belonged to the Gangetic belt of the eastern part of north India. The study was approved by ethics committee of the institute. Written consent was taken from all participants of the study. Patients suffering from leprosy, with bacteriological index (BI) ≥2 were included in the study. The samples (slit-skin smears) were obtained from patients before starting MDT, patients on MDT, defaulters of MDT (defined as who fail to complete treatment within the maximally allowed time frame i.e. six months treatment for PB leprosy must be completed within a maximum period of 9 months, similarly 12 months treatment for MB leprosy must be completed within 18 months), and patients who had relapse (defined as patients who developed new skin lesions after completion of MDT). Diagnosis was primarily on clinical grounds and confirmed by staining for acid fast bacilli (AFB) from samples obtained from ear lobes, forehead, or lesions on other sites of the body, by slit skin smear method, according to the standard procedures recommended by the WHO. Three smears were taken on a sterile glass slide and were air dried. One sample was stained by Ziehl–Neelsen staining and BI was calculated according to the Ridley's logarithmic scale.[18] Smears on the other two slides were scraped by a sterile blade and suspended in 1 ml sterile saline and stored in 1.5 ml air-tight micro-centrifuge tubes until processed in the department of microbiology.
Genomic DNA extraction
Samples were centrifuged at 10,000 revolutions per minute (rpm) for 5 min. Supernatants were discarded and pellets were re-suspended in 250 μl of tris-Cl EDTA-buffer (TE, pH 8.0) and mixed by vortexing. Then, 100 μl of 10% sodium dodecyl sulfate (SDS) and 3 μl of proteinase-K (20 mg/ml) were added and mixed by inverting the tubes. Cell suspensions were incubated at 37°C overnight. Subsequently, 100 μl of 5 M NaCl and 80 μl of 10% cetryl trimethyl ammonium bromide (C-TAB) were added to each tube and mixed well and incubated at 60°C for 10 min. Equal volume of phenol: chloroform: iso-amyl alcohol (IAA) (25:24:1) were added to the samples and centrifuged at 12,000 rpm for 10 min at 4°C to separate the aqueous and organic phase. Aqueous phase was collected in new tubes and mixed with equal volume of chloroform: IAA (24:1), and centrifuged at 12,000 rpm for 10 min at 4°C. Aqueous phase was collected again and mixed with equal volume of isopropanol and kept at room temperature for 10 min to precipitate the genomic DNA. Tubes were centrifuged at 12,000 rpm for 10 min at 4°C to precipitate the pellet. Supernatants were discarded and DNA pellets were washed with 70% ethanol by centrifugation at 8,000 rpm for 5 min. Pellets were dried at 37°C in inverted condition and dissolved in 50 μl TE.
Polymerase chain reaction amplification
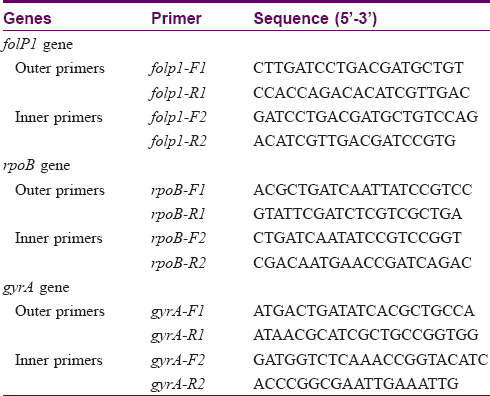
Each genomic DNA sample was submitted for primary and nested polymerase chain reaction (PCR) for amplification of drug resistance determining region of the rpoB, folP1, and gyrA genes with the primers used in earlier studies.[17] Primer sets folP1: F1/R1, rpoB: F1/R1 and gyrA: F1/R1 [Table - 1] were used for primary PCR to amplify the corresponding regions of respective genes. All the tubes were kept in thermocycler and standardized programs were used for PCR. For amplification of the target region of the folP1 gene, cycling conditions used were 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s for 35 cycles. For amplification of target regions of rpoB and gyrA genes, a programme of 30 s at 95°C, 30 s at 56°C, and 30 s at 72°C for 35 cycles were used. Primer sets [17]folP1: F2/R2, rpoB: F2/R2, and gyrA: F2/R2 [Table - 1] were used for the second round of amplification (nested PCR) using similar programs. Amplified products were resolved by agarose gel electrophoresis and visualized by ethidium bromide (EtBr) staining. Before submitting the amplified products for sequencing, every PCR product was purified using gel elution kit (Qiagen) according to the manufacturer's instructions. DNA sequencing was performed at Eurofin Genomics India Pvt. Ltd. (Bangalore, India) using chain-termination (Sanger's) method. DNA sequence analysis was done by pair-wise and multiple-sequence alignments using CLUSTAL W2 tool.
Results
Ninety-two patients were enrolled but only 78 (66 males and 12 females, mean age 35.73 ± 10.89 years) among them were included in the study. Remaining 14 patients'(5 before MDT, 6 on MDT, 2 defaulters and 1 relapse) smears failed to demonstrate M. leprae. The most common clinical type of leprosy was borderline lepromatous (41%) followed by lepromatous (38.5%), mid-borderline (15.4%), and histoid (5.1%). Primary amplification yielded the product of 254 base pair (bp), 345 bp, and 390 bp for folP1, rpoB, and gyrA genes, respectively. The secondary round of nested PCR yielded products of 255 bp, 242 bp, and 225 bp for rpoB, folP1, and gyrA, respectively. In 22 (28.2%) patients all the three genes could be amplified. In 39 (50%) cases rpoB, 32 (41.02%) folP1 and 45 (57.7%) cases gyrA were amplified. In 20 (25.6%) patients no gene could be amplified, in seven (8.9%) only rpoB, in four (5.1%) only folP1, and in 11 (14.1%) only gyrA were amplified. In 24 (30.8%) cases combination of rpoB and folP1, in 30 (38.5%) cases rpoB and gyrA and in 26 (33.3%) cases folP1 and gyrA were amplified by PCR. Forty five cases were new, 20 were on MDT, six were defaulters, and seven were relapsed cases. [Figure - 1],[Figure - 2],[Figure - 3] show PCR amplified products on agarose gel electrophoresis and visualized by ethidium bromide (EtBr) staining.
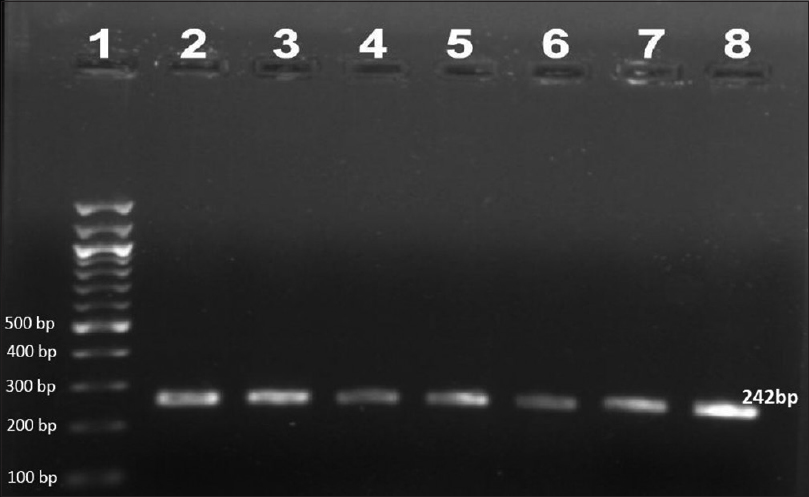 |
| Figure 1: Agarose gel electrophoresis of nested polymerase chain reaction product of the folP1 gene (242 bp)- lane 1–100 bp ladder, lane 2–8 folP1 gene product |
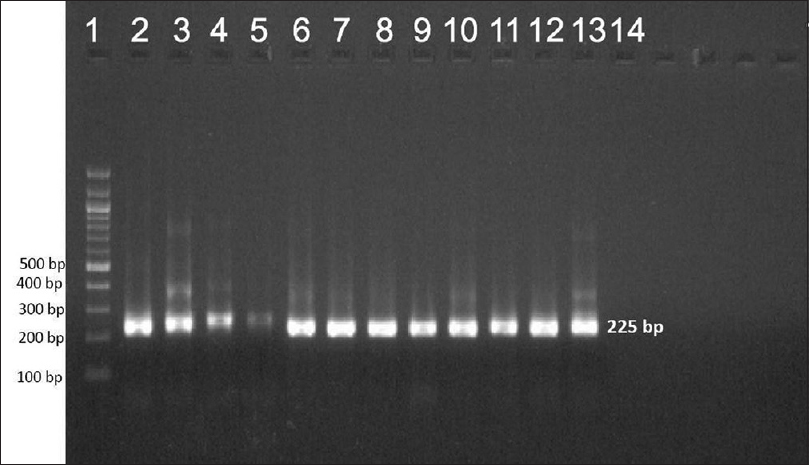 |
| Figure 2: Agarose gel electrophoresis of nested polymerase chain reaction product of the gyrA gene (225 bp)- lane 1–100 bp ladder, lane 2–13 gyrA gene product |
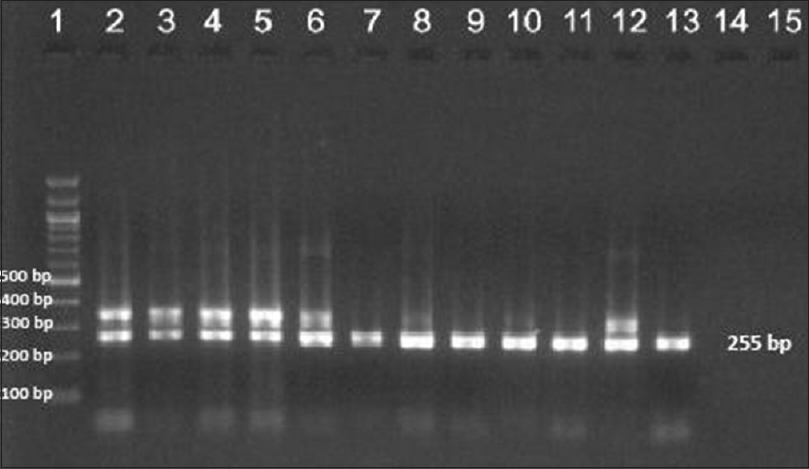 |
| Figure 3: Agarose gel electrophoresis of nested polymerase chain reaction product of the rpoB gene (255bp)- lane 1–100 bp ladder, lane 2–13 rpoB gene product |
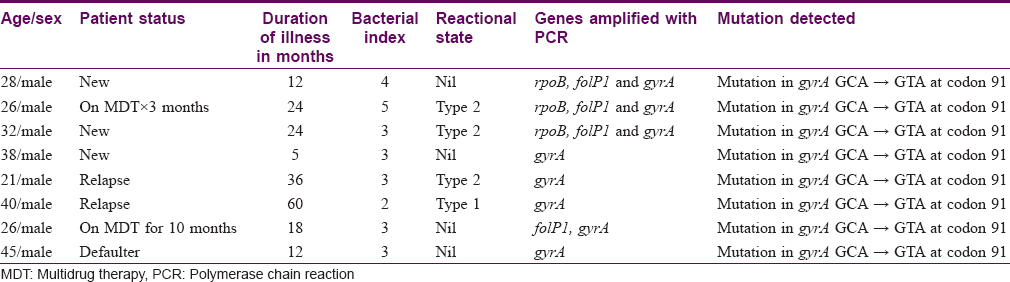
DNA sequencing of 32 samples of rpoB, 25 of folP1 and 38 of gyrA genes were successfully done. In the remaining samples, there were sequencing errors and therefore those cases were excluded from this analysis. Sequencing of rpoB genewas done on 22 new cases, six patients on MDT, two defaulters, and two cases of relapse. The expected mutation was not found in any of the samples. Sequencing of folP1 gene was done on 17 new cases, six patients on MDT and two defaulters. No mutation was found in any of them. Sequencing for gyrA gene was done on 24 new cases, seven patients on MDT, four defaulters and three cases of relapse. The expected mutations could be seen in only eight cases at 91 codon (GCA → GTA), of which three were new cases, two were on MDT, and three were defaulters. Details of patient characteristics with drug resistance are provided in [Table - 2].
Discussion
At present it is not technically possible to provide direct evidence for the mechanisms of resistance of M. leprae t o most antileprosy drugs. Current understanding regarding this aspect is based on our knowledge about drug resistance of M. tuberculosis.[19] According to several previous studies, drug resistance in M. leprae may be primarily attributed to mutations in genes encoding drug targets. For effective treatment and containment of drug resistant strains, it is mandatory to have local and global data on the drug resistance pattern of the bacterium. In the present study, we have examined the mutations possibly associated with drug resistance in the target genes by PCR-based amplification and sequencing. It is really heartening to note that none of the randomly selected 32 amplicons subjected for sequencing showed nucleotide mutation in rpoB, indicating that the M. leprae strains on eastern part of North India are 100% sensitive to rifampicin. Our finding is in agreement with another study carried out in South India in 2011 by Sekar et al.[20] On the contrary, one study conducted on relapse cases of leprosy, collected from leprosy hospitals scattered wide across regions endemic for leprosy in India, during 2009 and 2013, showed mutations in rpoB gene of the M. leprae in 3.6% (4/111) at codon 439 (Phe → Leu), at 442 (Gln → His), 433 (Thr → Ile), and at 441 (Asp → Tyr).[21] Another study carried out in east India showed 4% (2/50) mutation at codon 442 (Glu → His) in relapse cases.[22] The latter mutation (Glu → His) was common with other Indian studies. There was no sharing with those 4 mutations reported from east and southeast Asian countries in strains of relapsed cases.[17],[23]
The folP1 gene, a target for dapsone, showed no mutation, indicating that dapsone is likely to be very effective in treating patients with leprosy in this region. However, previous studies from other parts of India had shown that the frequency of mutations of M. leprae among relapsed cases ranged from 8.1% to 15%.[20],[21]
While looking for mutations in the target gene gyrA for quinolone, we found that 21% (8/38) of the strains had only one point mutation i.e. at 91 codon, from GCA → GTA leading to change in amino acid from alanine → valine. These eight cases consisted of three new cases, two patients on MDT, one defaulter, and two cases of relapse. The finding of this mutation in three new cases in our study was surprising. A possible reason is rampant prescription of quinolones to treat other infections. A study carried out on patients attached to Leprosy Mission hospitals in India showed mutations in gyrA gene in 8.1% (9/111) of the participants.[21] Three different mutations were noted in this study; two at codon 91 (Ala → Thr); 91 (Ala → Val), and one at codon 92 (Ser → Ala). However, Sekar et al. from south India did not find any mutation in this gene.[20] A study conducted in east and southeast Asian countries showed mutations in gyrA gene among 6.8% of the participants.[23] A study from Japan found only one strain with mutation at codon 91 (Ala → Val).[12] Surprisingly, several studies carried out in different parts of the world (India, USA, Japan, Vietnam, etc.) reported no mutation in the gyrA gene of M. leprae.[17],[24],[25] Our region is endemic for enteric fever also and quinolones were most commonly used to treat these infections. Quinolones were misused to treat many other types of infections and febrile conditions which may lead to drug resistance with these drugs. A high rate of mutation in gyrA gene of M. leprae from this region indicates the weak prospects of quinolone as a second line drug.
Small number of cases and not focussing on relapsed cases are possible limitations of this study. Our findings indicate that resistance to first line drugs against leprosy is not a major problem in east Uttar Pradesh. We must counsel the patients about adherence to treatment and follow-up the patients adequately to minimize the number of defaulters. However, it must also be noted that mutations of the three genes rpoB, folP1, and gyrA must not be taken as a definite indicator of drug resistance. The culprit mutation must be verified by mouse foot pad assay meticulously. Further, such mutations must also be examined in strains cured later by standard therapy.
To conclude: Antileprotic drugs present in MDT are still effective against Mycobacterium leprae. Quinolones may not be a good choice as an antileprotic agent in our region.
Acknowledgement
We are thankful toAjita and Shailendra Singh for their technical support in laboratory.
Financial support and sponsorship
Institutional research grant from Banaras Hindu University, Varanasi.
Conflicts of interest
There are no conflicts of interest.
| 1. |
World Health Organization. Global leprosy update. Wkly Epidemiol Rec 2014;89:389-400.
[Google Scholar]
|
| 2. |
Desikan KV. Elimination of leprosy and possibility of eradication-The Indian scenario. Indian J Med Res 2012;135:3-5.
[Google Scholar]
|
| 3. |
Pettit JH, Rees RJ. Sulphone resistance in leprosy. An experimental and clinical study. Lancet 1964;2:673-4.
[Google Scholar]
|
| 4. |
Pearson JM, Rees RJ, Waters MF. Sulphone resistance in leprosy. A review of one hundred proven clinical cases. Lancet 1975;2:69-72.
[Google Scholar]
|
| 5. |
Grosset JH, Guelpa-Lauras CC, Bobin P, Brucker G, Cartel JL, Constant-Desportes M, et al. Study of 39 documented relapses of multibacillary leprosy after treatment with rifampin. Int J Lepr Other Mycobact Dis 1989;57:607-14.
[Google Scholar]
|
| 6. |
Jacobson RR, Hastings RC. Rifampin-resistant leprosy. Lancet 1976;2:1304-5.
[Google Scholar]
|
| 7. |
Ji B, Perani EG, Petinom C, Grosset JH. Bactericidal activities of combinations of new drugs against Mycobacterium leprae in nude mice. Antimicrob Agents Chemother 1996;40:393-9.
[Google Scholar]
|
| 8. |
Shepard CC, Chang YT. Effect of several anti-leprosy drugs on multiplication of human leprosy bacilli in footpads of mice. Proc Soc Exp Biol Med 1962;109:636-8.
[Google Scholar]
|
| 9. |
Cole ST, Eiglmeier K, Parkhill J, James KD, Thomson NR, Wheeler PR, et al. Massive gene decay in the leprosy bacillus. Nature 2001;409:1007-11.
[Google Scholar]
|
| 10. |
Eiglmeier K, Parkhill J, Honoré N, Garnier T, Tekaia F, Telenti A, et al. The decaying genome of Mycobacterium leprae. Lepr Rev 2001;72:387-98.
[Google Scholar]
|
| 11. |
Scollard DM, Adams LB, Gillis TP, Krahenbuhl JL, Truman RW, Williams DL. The continuing challenges of leprosy. Clin Microbiol Rev 2006;19:338-81.
[Google Scholar]
|
| 12. |
Matsuoka M, Suzuki Y, Garcia IE, Fafutis-Morris M, Vargas-González A, Carreño-Martinez C, et al. Possible mode of emergence for drug-resistant leprosy is revealed by an analysis of samples from Mexico. Jpn J Infect Dis 2010;63:412-6.
[Google Scholar]
|
| 13. |
Williums DL, Spring L, Harris E, Roche P, Gills TP. Dihydropteroate Synthase of Mycobacterium leprae and Dapsone Resistance. Antimicrob Agents Chemother. 2000; 44 (6): 1530–1537.
[Google Scholar]
|
| 14. |
Menzies D, Benedetti A, Paydar A, Martin I, Royce S, Pai M, et al. Effect of duration and intermittency of rifampin on tuberculosis treatment outcomes: A systematic review and meta-analysis. PLoS Med 2009;6:e1000146.
[Google Scholar]
|
| 15. |
Jamet P, Ji B. Relapse after long-term follow up of multibacillary patients treated by WHO multidrug regimen. Marchoux Chemotherapy Study Group. Int J Lepr Other Mycobact Dis 1995;63:195-201.
[Google Scholar]
|
| 16. |
Chen XS, Li WZ, Jiang C, Ye GY. Studies on risk of leprosy relapses in China: relapses after treatment with multidrug therapy. Int J Lepr Other Mycobact Dis 1999;67:379-87.
[Google Scholar]
|
| 17. |
Matsuoka M, Budiawan T, Aye KS, Kyaw K, Tan EV, Cruz ED, et al. The frequency of drug resistance mutations in Mycobacterium leprae isolates in untreated and relapsed leprosy patients from Myanmar, Indonesia and the Philippines. Lepr Rev 2007;78:343-52.
[Google Scholar]
|
| 18. |
Ridley DS, Jopling WH. Classification of leprosy according to immunity. A five-group system. Int J Lepr Other Mycobact Dis 1966;34:255-73.
[Google Scholar]
|
| 19. |
Musser JM. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin Microbiol Rev 1995;8:496-514.
[Google Scholar]
|
| 20. |
Sekar B, Arunagiri K, Kumar BN, Narayanan S, Menaka K, Oommen PK. Detection of mutations in folp1, rpoB and gyrA genes of M. leprae by PCR-direct sequencing – A rapid tool for screening drug resistance in leprosy. Lepr Rev 2011;82:36-45.
[Google Scholar]
|
| 21. |
Lavania M, Jadhav RS, Chaitanya VS, Turankar R, Selvasekhar A, Das L, et al. Drug resistance patterns in Mycobacterium leprae isolates from relapsed leprosy patients attending The Leprosy Mission (TLM) Hospitals in India. Lepr Rev 2014;85:177-85.
[Google Scholar]
|
| 22. |
Hasanoor Reja AH, Biswas N, Biswas S, Lavania M, Chaitanya VS, Banerjee S, et al. Report of rpoB mutation in clinically suspected cases of drug resistant leprosy: A study from Eastern India. Indian J Dermatol Venereol Leprol 2015;81:155-61.
[Google Scholar]
|
| 23. |
Maeda S, Matsuoka M, Nakata N, Kai M, Maeda Y, Hashimoto K, et al. Multidrug resistant Mycobacterium leprae from patients with leprosy. Antimicrob Agents Chemother 2001;45:3635-9.
[Google Scholar]
|
| 24. |
Kai M, Nguyen Phuc NH, Nguyen HA, Pham TH, Nguyen KH, Miyamoto Y, et al. Analysis of drug-resistant strains of Mycobacterium leprae in an endemic area of Vietnam. Clin Infect Dis 2011;52:e127-32.
[Google Scholar]
|
| 25. |
Williams DL, Spring L, Harris E, Roche P, Gillis TP. Dihydropteroate synthase of Mycobacterium leprae and dapsone resistance. Antimicrob Agents Chemother 2000;44:1530-7.
[Google Scholar]
|
Fulltext Views
6,294
PDF downloads
3,148





