Translate this page into:
A novel loci of the HR gene in Marie - Unna hereditary hypotrichosis using whole-exome sequencing
2 Catholic Precision Medicine Research Center, College of Medicine, The Catholic University of Korea, Seoul, Korea
3 Department of Dermatology, College of Medicine, The Catholic University of Korea, Seoul, Korea
Correspondence Address:
Young Bok Lee
Department of Dermatology, Uijeongbu St. Mary's Hospital, College of Medicine, The Catholic University of Korea, 271 Chunbo Street, Uijeongbu, 07345
Korea
| How to cite this article: Lee M, Lee G, Chung YJ, Kang MJ, Yu DS, Lee YB. A novel loci of the HR gene in Marie - Unna hereditary hypotrichosis using whole-exome sequencing. Indian J Dermatol Venereol Leprol 2020;86:321-324 |
Sir,
Marie - Unna hereditary hypotrichosis is a rare and distinctive form of congenital hypotrichosis. It is a rare autosomal dominant hair disorder with the notable absence of non-dermatological signs that are typical components of syndromic hereditary hypotrichosis, such as retinal degeneration, hearing impairment or intellectual disability. Patients with Marie - Unna hereditary hypotrichosis are usually born with sparse hair but later regrowth of coarse hair occurs. As they age, hair loss gradually occurs on the scalp, eyebrows and eyelashes and progresses to complete baldness.
The human hairless [HR] gene has been reported to cause autosomal dominant Marie - Unna hereditary hypotrichosis (MIM 146550), autosomal recessive atrichia with papular lesions (MIM 209500) and alopecia universalis congenita (MIM 203655). The HR gene spans over 14kb on chromosome 8p21 and is composed of 19 exons. The protein product of the HR gene has a zinc-finger domain and is a putative transcription co-repressor for multiple nuclear receptors that are highly expressed in the brain, epidermis and hair follicles.[1] In Marie - Unna hereditary hypotrichosis, genetic mutations have been reported in upstream open reading frames in the 5' untranslated region of HR in several studies.[2],[3],[4] Mutations in U2HR (second open reading frame in the 5' untranslated region) increase expression of the HR gene and lead to disruption in hair follicle morphogenesis and cycling.[2] Until now, there have been no reports of genetic variants located in the protein-coding regions of HR.
A 21-year-old male presented with progressive alopecia on the scalp that began in childhood. He had patches of alopecia on the scalp, particularly the occipital margins, with coarse hair on the frontal area. Eyebrows and eyelashes were absent but there were no ectodermal abnormalities. Laboratory findings were normal. Histological examination revealed a reduced number of hair follicles; however, inflammation was not present around the hair follicles. Microscopic examination of the hair shafts did not reveal the abnormalities typical of fragile hair shaft disorders, including trichorrhexis nodosa, trichorrhexis invaginata, pili torti or monilethrix. The patient reported that his father (49 years old) also had complete baldness that began in childhood and his 9-year-old sister had been exhibiting progressive alopecia for the past several years [Figure - 1]. After obtaining approval from the Institutional Review Board at Uijeongbu St. Mary's Hospital, Catholic University of Korea (UC16TISI0167) and informed consent from the patient and family members, blood samples were collected and kept in Uijeongbu St. Mary's Hospital Human Biobank. The blood of the three affected patients and family members without hair loss (n = 4) were provided for whole-exome sequencing. Whole exome sequencing was performed on an Illumina HiSeq 2500 system (Illumina, San Diego, CA, USA). Sequenced reads were aligned to a reference genome using the Burrows–Wheeler Aligner. The average mapping depth for each sample was larger than 110-fold. Raw variants were called using GATK4, and low quality (Q-score < 20) and low depth (<10-fold) variants were filtered out. In addition, variants with global minor allele frequencies greater than 0.001 in either all populations or the East Asian population of the Genome Aggregation Database were excluded. Finally, we identified six single nucleotide variants satisfying an autosomal dominant pattern in exonic regions. Of the six single nucleotide variants, three were nonsynonymous and the other three were synonymous single nucleotide variants [Table - 1]. No insertions, deletions or frameshift variants were identified. For the three nonsynonymous single nucleotide variants (HASPIN, NLRP8, and ATAD3), the pathogenic effect was predicted using Sorting Intolerant from Tolerant (SIFT) and PolyPhen. However, we could not find any evidence for the relationship between the single nucleotide variants/their genes and hair loss or related symptoms via literature search and “Open Targets Platform”[5] (is a web application that provides an interface to the data that is being aggregated and reprocessed on a regular basis from an increasing list of valuable public data sources). Among the synonymous single nucleotide variants, one variant was located in exon 2 of HR [Figure - 2]a. We carried out Sanger sequencing for the six single nucleotide variants identified on whole-exome sequencing and all genotypes matched in the results from both methods [Figure - 2]b.
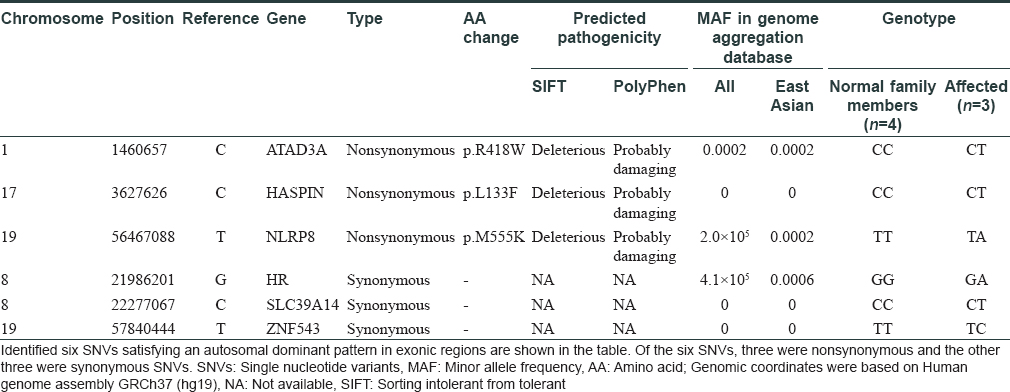
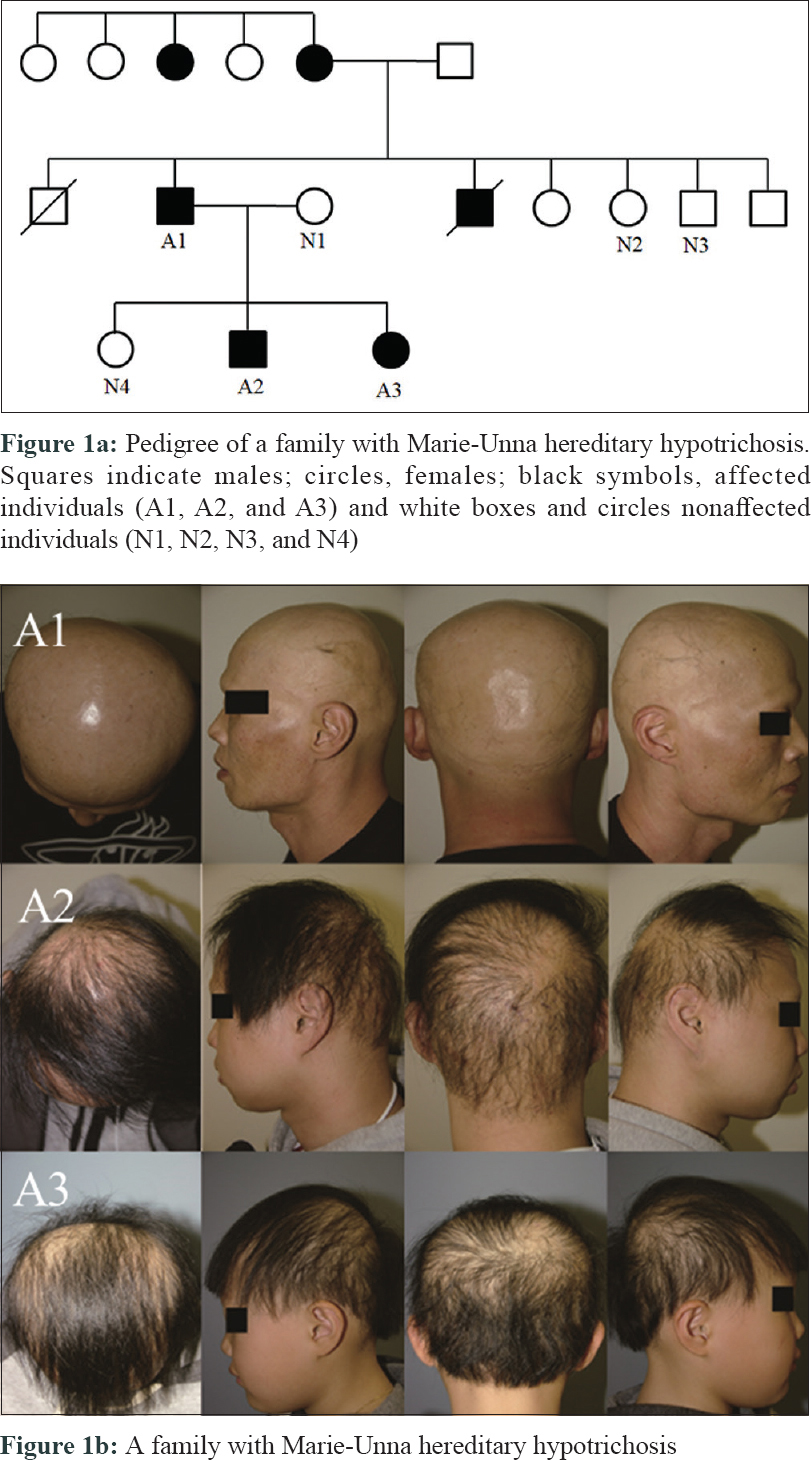 |
| Figure 1: |
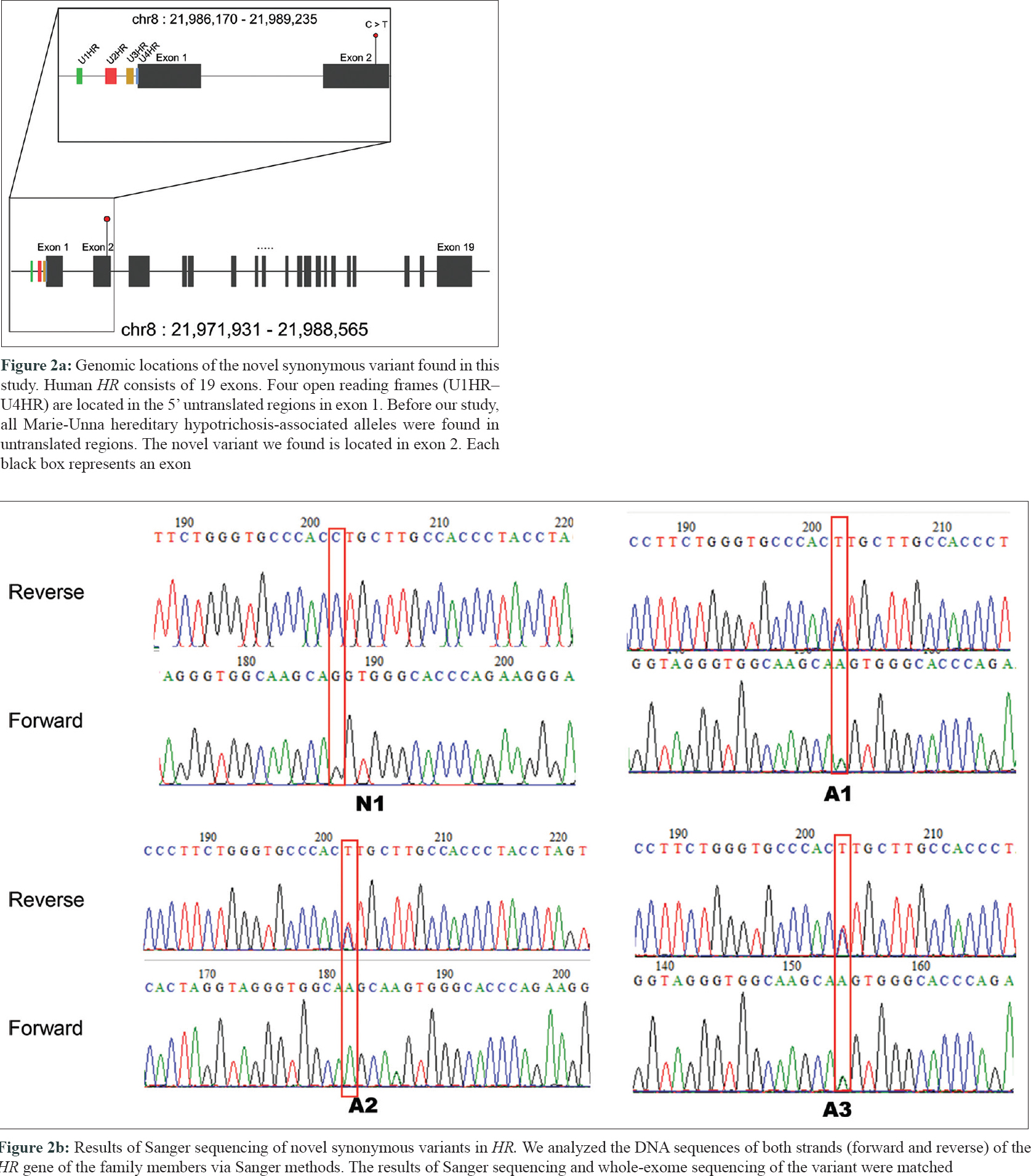 |
| Figure 2: |
Until now, heterozygous autosomal dominant mutations in the U2HR frame of the HR gene have been reported in families with Marie - Unna hereditary hypotrichosis worldwide.[2],[3],[4] However, we could not find mutations in the U2HR in the affected individuals. Although whole-exome sequencing was performed to find a disease-causing mutation within the coding regions with a high sequencing depth (110×), we could detect a disease-causing mutation on the untranslated regions that are sequenced in the process of exome target enrichment.[6] Previously reported mutations in U2HR caused a change in RNA expression of HR without changing the amino acid sequence of HR.[2] The possible molecular mechanism of Marie - Unna hereditary hypotrichosis is gain-of-function of the HR.[2] Recent studies have shown that synonymous variants can change protein folding and function as well as mRNA structure and splicing. If the synonymous single nucleotide variant leads to altered mRNA and protein expression of HR, a novel molecular cis-regulatory mechanism for HR can be established. Although it was a synonymous single nucleotide variant, determining its role in Marie - Unna hereditary hypotrichosis will require further validation. But based on the previous functional studies of the HR gene, we suggest that this variant in HR may be a genetic factor in this affected family. Further mouse model studies to validate this association and establish molecular mechanisms on how synonymous mutations could cause hair loss have to be done.
Declaration of patient consent
The authors certify that they have obtained all appropriate patient consent forms. In the form, the patient has given his consent for his images and other clinical information to be reported in the journal. The patient understands that name and initials will not be published and due efforts will be made to conceal identity, but anonymity cannot be guaranteed.
Financial support and sponsorship
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government Ministry of Science and Information and Communication Technology (MSIP) (NRF-2019R1F1A1056601).
Conflicts of interest
There are no conflicts of interest.
| 1. |
Cachon-Gonzalez MB, Fenner S, Coffin JM, Moran C, Best S, Stoye JP. Structure and expression of the hairless gene of mice. Proc Natl Acad Sci U S A 1994;91:7717-21.
[Google Scholar]
|
| 2. |
Wen Y, Liu Y, Xu Y, Zhao Y, Hua R, Wang K, et al. Loss-of-function mutations of an inhibitory upstream ORF in the human hairless transcript cause Marie-Unna hereditary hypotrichosis. Nat Genet 2009;41:228-33.
[Google Scholar]
|
| 3. |
Redler S, Kruse R, Eigelshoven S, Hanneken S, Refke M, Wen Y, et al. Marie Unna hereditary hypotrichosis: Identification of a U2HR mutation in the family from the original 1925 report. J Am Acad Dermatol 2011;64:e45-50.
[Google Scholar]
|
| 4. |
Yang J, Liang Y, Zeng K, Huang L, Zheng M. Marie Unna hereditary hypotrichosis: A recurrent c. 74C and T mutation in the U2HR gene and literature review. Int J Dermatol 2014;53:206-9.
[Google Scholar]
|
| 5. |
Koscielny G, An P, Carvalho-Silva D, Cham JA, Fumis L, Gasparyan R, et al. Open Targets: A platform for therapeutic target identification and validation. Nucleic Acids Res 2017;45:D985-94.
[Google Scholar]
|
| 6. |
Samuels DC, Han L, Li J, Quanghu S, Clark TA, Shyr Y, et al. Finding the lost treasures in exome sequencing data. Trends Genet 2013;29:593-9.
[Google Scholar]
|
Fulltext Views
4,680
PDF downloads
2,527





