Translate this page into:
Association between human leukocyte antigen-DRB1 and human leukocyte antigen-DQB1 alleles and pemphigus vulgaris in Indian patients: A case–control study
2 Department of Transfusion Medicine and Immunohematology, Christian Medical College, Vellore, Tamil Nadu, India
3 Department of Nephrology, Christian Medical College, Vellore, Tamil Nadu, India
4 Department of Biostatistics, Christian Medical College, Vellore, Tamil Nadu, India
Correspondence Address:
Renu George
Department of Dermatology, Christian Medical College Ida Scudder Road, Vellore - 632 004, Tamil Nadu
India
| How to cite this article: Priyadarshini A, George R, Daniel D, Varughese S, Jayaseelan V. Association between human leukocyte antigen-DRB1 and human leukocyte antigen-DQB1 alleles and pemphigus vulgaris in Indian patients: A case–control study. Indian J Dermatol Venereol Leprol 2018;84:280-284 |
Abstract
Background: HLA-DRB1*04, -DRB1*08, -DRB1*14, -DQB1*03 and -DQB1*05 are reported to have significant association with pemphigus vulgaris; however, this is partially dependent on ethnicity. This study was done to determine the HLA-DR and -DQ types prevalent in Indian patients with pemphigus vulgaris.
Methods: A prospective case–control study was done for a period of 9 months in Christian Medical College Vellore, India. HLA typing was done by PCR-SSOP method in 50 cases and 50 healthy controls. Allele frequencies in cases and controls were compared and odds ratios with 95% confidence interval were calculated.
Results: The mean age of the patients (29 females, 21 males) and that of controls (36 males, 14 females) were 41.3 ± 13.65 and 35.42 ± 11.09 years, respectively. HLA-DRB1*14 was present in 47 patients and 18 controls (OR, 27.85; 95% CI, 7.57–102.42) and HLA-DQB1*05 was seen in 47 patients and 24 controls (OR, 16.97; 95% CI, 4.66–61.80). The haplotype DRB1*14, DQB1*05 was present in 44 patients and 14 controls (OR, 18.86; 95% CI, 6.58–54.05). DRB1*15 was present in 7 cases and 16 controls (OR, 0.35; 95% CI, 0.13–0.94) and DQB1*06 was present in 8 cases and 19 controls (OR, 0.31; 95% CI, 0.12–0.80). HLA-DQB1*03 was associated with significantly higher pemphigus disease area index scores.
Limitations: The main limitations were that the numbers studied were small as the study was conducted at a single center, and the haplotype analysis was limited only to the proband. PDAI scores could have been influenced by prior treatment.
Conclusion: There was a significant association between HLA-DRB1*14 and HLA-DQB1*05 and pemphigus vulgaris in our patients. A negative association was seen with DRB1*15 and DQB1*06.
Introduction
Pemphigus is a group of chronic autoimmune bullous diseases that involves the skin and mucosa. It has a worldwide prevalence varying from 0.76 to 5 per million.[1] The disease is more prevalent in India and Middle East than in Western Europe or North America.[2] It is broadly divided into four types: pemphigus vulgaris (PV), pemphigus foliaceus, paraneoplastic pemphigus and drug-induced pemphigus. The disease results from the interplay between genetic and environmental factors. Genetic susceptibility to pemphigus vulgaris has been demonstrated by allele and haplotype studies from different parts of the world.[3] It is hypothesized that distinct HLA class II alleles shape the autoimmune response to desmoglein 3.[4]
Over 95% pemphigus patients have specific HLA-DRB1 or DQB1 or both alleles.[3] Of the DRB1 alleles nine subtypes, DRB1*0402, *0403, *0406, *0802, *0804, *1401, *1404, *1405 and *1408, have been associated with PV as reported from various studies on different population groups.[4],[5],[6],[7],[8],[9],[10] Of these, DRB1*0402 is more common in the European and American patients with PV, while DRB1*1401 and DRB1*1404 are more common in the Indo-Asian pemphigus patients.[11] DQB1*0302 and DQB1*0503 are also reported worldwide in patients with PV. DQB1*0302 is classically associated with PV in Ashkenazi Jews, while DQB1*0503 has been reported from other European, Japanese and Pakistani patients.[6],[7],[9],[10],[11],[12],[13]
Thus there is significant variation in the HLA allele types that have been reported in association with PV from different ethnic populations around the world. There is, however, a paucity of data in this field from India. Therefore, the primary objective of the study was to determine the HLA-DR and DQ types prevalent in Indian patients with PV. The secondary objective was to determine the correlation between the prevalent HLA type and disease activity assessed by pemphigus disease area index (PDAI) score in patients with active disease.
Methods
The study was a hospital-based case–control study of patients with PV either admitted or attending the dermatology outpatient department, Christian Medical College, Vellore, Tamil Nadu, India. It was conducted between November 2013 and August 2014 for a period of 9 months. The study was approved by the Institutional Review Board (IRB approval no. 8549).
The inclusion criteria for cases were patients with PV diagnosed by clinical features, histopathology and immunofluorescence or serological demonstration of anti-desmoglein antibodies and consenting for the study. Healthy renal transplant donors who were willing to participate in the study were chosen as controls. Sample size was calculated to be 45 cases and 45 controls. It was calculated to show a difference in HLA allele of about 30% across patients and controls, 80% of the time with 5% level of significance, based on the prevalence data of previous studies.[14] Detailed history and clinical examination findings were recorded. Disease severity was assessed by PDAI.[15] Low-resolution HLA-DRB1 and DQB1 typing was done by PCR using sequence-specific oligonucleotide probes for patients and healthy controls.
The data were entered in EpiData version 3.1, and analyzed using the software SPSS version 17. The HLA allele frequencies in cases and controls were compared and odds ratios and 95% confidence interval were calculated. The Kruskal–Wallis test was used to analyze the relation between HLA-DR and -DQ types with disease severity.
Results
Fifty-four patients with pemphigus vulgaris (32 females and 22 males) of Indian origin were seen during the study period. HLA typing was done for 50 cases. The last four patients were excluded from HLA testing due to lack of funds.
Demographic and clinical characteristics of patients and controls
The demographic and clinical characteristics of the patients included in the study are shown in [Table - 1]. All the patients were from India, majority being from Tamil Nadu. The mean age of the patients was 41.3 ± 13.65 years (range, 12–66 years). The mean age at the onset of disease was 38.47 ± 13.11 years (range, 12–61 years). Four patients developed the disease before 20 years of age. There was slight female predominance with 29 females and 21 males; M:F ratio 1:1.38. At the time of enrolment, 40/50 patients had active disease. The mean PDAI was 23.9. None of the patients had relatives with pemphigus.
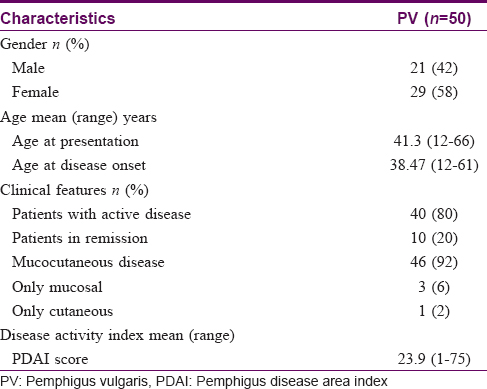
Fifty healthy renal transplant donors who were willing to participate in the study were chosen as controls. The mean age of the controls (36 males and 14 females) was 35.42 ± 11.09 years (range, 20–59 years).
HLA-DRB1 and -DQB1 alleles and haplotypes frequency distribution in pemphigus vulgaris patients as compared to healthy controls
The frequency distribution of HLA-DRB1 and -DQB1 alleles and haplotypes in patients and controls is shown in [Table - 2]. The common allele types and haplotypes were distributed evenly in males and females; there was no statistically significant difference based on gender. DRB1*14 was present in 47 patients and 18 controls (OR: 27.85; 95%CI: 7.57, 102.42) (P< 0.001). Nine patients and one control were homozygous for the allele. HLA-DQB1*05 was present in 47 patients and 24 controls (OR: 16.97; 95%CI: 4.66, 61.80) (P< 0.001). Ten patients were homozygous for the allele. Haplotype analysis revealed that 44/50 patients had the haplotype DRB1*14, DQB1*05 versus 14/50 controls (OR:18.86; 95% CI: 6.58, 54.05) (P< 0.001).
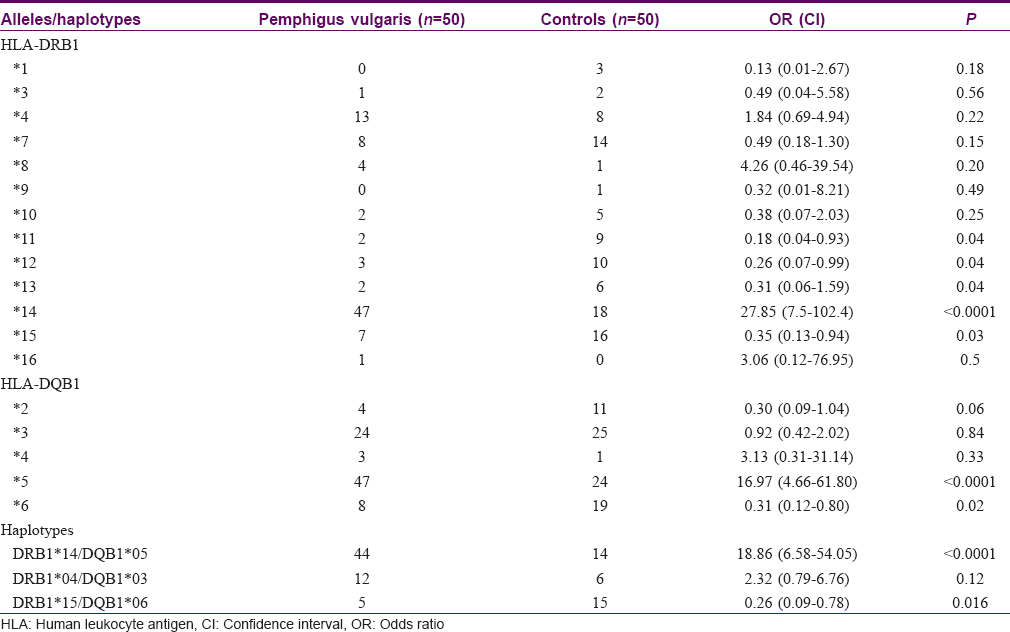
The allele DRB1*15 was present in 7 cases versus 16 controls (OR: 0.35; 95% CI: 0.13, 0.94) (P = 0.03) and DQB1*06 was present in 8 cases versus 19 controls (OR: 0.31; 95% CI: 0.12, 0.80) (P = 0.02). The other HLA alleles that were found to have a significantly higher prevalence in controls were DRB1*11, DRB1*12, DRB1*13, but the numbers studied were small.
Association between HLA -DRB1, DQB1 alleles, haplotypes, and PDAI
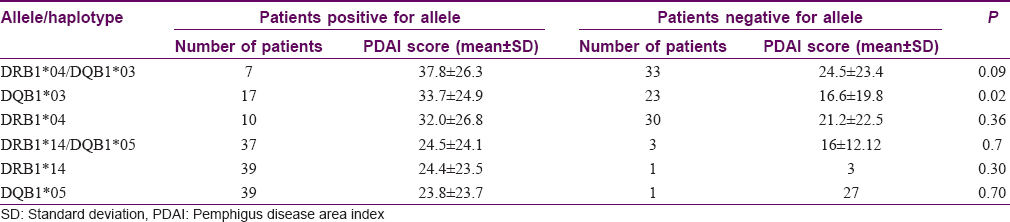
During the study period, 40/50 patients presented with active disease. The mean PDAI score was 23.9 (range 1–75). At the time of inclusion of this study, 37/40 patients were on treatment. All of these 37 patients were on corticosteroids. The most common adjuvant was azathioprine in 28 patients. The presence of HLA-DQB1*03 (in 17/40 cases) was associated with significantly higher PDAI scores (mean score = 33.7) as compared to that of patients with absence of the allele (23/40 cases, mean PDAI = 16.6) with P value of 0.02. However, all 17/40 patients with HLA-DQB1*03 also had DQB1*05 and all but one had DRB1*14 [Table - 3].
Discussion
The role of genes in the predisposition to pemphigus has been repeatedly reinforced by numerous studies done worldwide. The female predominance in PV, as seen in this study, has been reported in various studies from India and across the globe.[16],[17] This is similar to other diseases of autoimmune etiology and is probably a hint to the role of genetics in the pathogenesis of PV.[3] Studies demonstrate significantly higher frequency of occurrence of certain MHC class II genes in patients with pemphigus as compared to healthy individuals. In a country like India, where PV is not a rare disease, the data regarding HLA gene association is limited.[11],[14],[18] Two of the three studies were published two decades ago. This single-center case–control study from India provides robust data on the association between HLA-DRB1 and HLA-DQB1 alleles and pemphigus vulgaris in Indian patients.
We found a significant association of DRB1*14 with PV in our study. This association of HLA-DRB1*14 with PV has been supported by a meta-analysis that included 18 studies from different parts of the world including Indian patients, as well as previous study from India.[5],[14] This particular allele is more often associated with patients of Asian origin as compared to the European Caucasian population which is in concurrence with our data.[11] Although the DRB1*04 allele, the most prevalent allele in Ashkenazi Jews, Caucasians, Iranian and Japanese pemphigus patients, was present in 26% of patients, the association did not prove to be significant. This finding again was similar to previous study on Indian patients.[14] The DRB1*04 allele has also been reported as the significant allele associated with PV from Pakistani patients.[19] The increased frequency of HLA-DRB1*15 in controls versus patients with PV that was seen in our study was also noted by other studies from the West (but not from India), suggesting a protective role in pemphigus.[5],[6],[9]
Among the different DQB alleles, we found a significant association of PV with DQB1*05. Our result is consistent with other studies from India and from other parts of the world that have looked at DQB1 alleles in pemphigus.[6],[7],[9],[10],[11],[12],[14] HLA-DQB1*03 was not found to be of significant association in our study, which is in concordance with most other studies on Indian and Asian patients, but this particular allele is of significance in European and Caucasian pemphigus patients.[6],[9],[12] It was, however, found to be significant in a subset of patients in a single study from India.[14] Significant negative association with DQB1*06 was also noted in our study as has been reported from other studies.[6],[13]
With respect to haplotype analysis, we found a strong association for DRB1*14, DQB1*05. Our result is consistent with that of other studies that have analyzed haplotypes.[6],[7],[10],[12],[14] It is of interest to note that the haplotype DRB1*04, DQB1*03 that has been identified as the characteristic haplotype contributing to the development of pemphigus in Ashkenazi Jews was not statistically significant in our study. Studies that have included the analysis of DQA1 alleles have found the haplotype DRB1*04, DQB1*03, DQA1*03 to be significantly associated with pemphigus patients of Ashkenazi Jewish and non-Jewish Caucasian patients, and DRB1*14, DQB1*05, DQA1*01 to be associated with pemphigus patients of Middle-Eastern countries and Pakistan.[6],[7],[12]
The HLA genes exhibit a high degree of polymorphism. The polymorphism for the class II genes occurs due to change in nucleotide sequences coding for alpha-1 and beta-1 domains that form the peptide binding pocket. Variation in the peptide binding pocket that occurs due to expression of different alleles is responsible for change in antigen presentation and consequently is of paramount importance in causation of autoimmune disease.[20] The difference between the peptide binding pocket of disease-associated HLA-DR and DQ types in comparison with their normal types explains the structural basis by which specific HLA types confer susceptibility to pemphigus. The presence of negatively charged amino acids in the beta-chain at the peptide binding site is responsible for the binding of the positively charged desmogleins peptides which are the main target antigens of pemphigus.[20] This specificity of peptide binding pocket has been seen in DRB1*04 and DQB1*05. It has been therefore suggested that DQB1*03 and DRB1*14 are found in higher frequency only by virtue of linkage disequilibrium with the aforementioned alleles, respectively.[6],[7],[12],[21] If this is considered then the allele contributing to the development of pemphigus in Indian patients would be primarily HLA-DQB1*05. The role of DQA1 alleles has, however, not yet been investigated further.
The secondary objective of this study was to determine if there was any correlation between the prevalent HLA type and disease activity assessed by PDAI score in patients with active disease, as has been reported previously.[22] The allele DQB1*03 was associated with more severe disease as seen by higher PDAI scores in patients carrying this allele. This finding was similar to the results of a recent study wherein DRB1*04, DRB1*14 and DQB1*03, DQB1*05 alleles were correlated with disease severity (as measured by ABSIS score).[22] They also reported an association for DRB1*04 and severe disease, which was not seen in our study. However, in the present study in patients with active disease, the DQB1*03 allele was associated with DQB1*05 in all the cases and with DRB1*14 in all except one case. Therefore, no definitive conclusion can be drawn regarding its predictability for more severe disease.
The present study highlights the association of alleles HLA-DRB1*14 and -DQB1*05 and the haplotype DRB1*14, DQB1*05 with pemphigus patients in India. The sample size was calculated for the presence of HLA-DRB1 and -DQB1 across the cases and controls. It is evident from the tables provided that the number was sufficient enough as the association of some of the alleles with PV were statistically significant. This is, however, a single-center study, hence the numbers studied are small. Large multicenter studies will provide more comprehensive data on the HLA associations of pemphigus in Indian patients including that of pemphigus foliaceus. Other limitations were that subtyping of HLA alleles was not available and HLA-DQA types were not studied. Also, the haplotype analysis was limited only to the proband; the HLA-DR, DQ types of parents were not looked at. We studied the correlation between the HLA type and PDAI in patients who had active disease; however, these patients were on treatment which may have influenced the PDAI score.
Further insight into the genetic basis for pemphigus may come from studying the HLA and non-HLA genes in pemphigus patients in comparison with the close relatives. The non-HLA genes that have been identified by candidate-driven gene studies and genome-wide studies include protein tyrosine phosphatase N22 (PTPN22), single nucleotide polymorphisms in ST18 gene and desmoglein 3, particularly DSG3*TCCCC was found to be of significance in the Indian pemphigus patients.[3],[23],[24] The association of alleles with disease severity could also be assessed by prospective multicenter studies. Targeted blockade of PV-associated HLA alleles by topically applied proteomics derived desmoglein peptide has resulted in significant clinical improvement as well as a decrease in anti-Dsg antibody titre.[25] Thus studies on PV-associated HLA alleles will not only result in a better understanding of the genetic basis of the disease but may also provide data that may help in evolving better treatment modalities for pemphigus patients.
Conclusion
The present study has shown that there is a significant association between HLA-DRB1*14, -DQB1*05 and the haplotype DRB1*14, DQB1*05 and pemphigus vulgaris in Indian patients. On the other hand, HLA-DRB1*15 and -DQB1*06 showed a negative association with PV. As there is limited data on the association between pemphigus vulgaris and HLA alleles from India, large multicenter studies are required for more comprehensive data on the association between HLA alleles and the different types of pemphigus.
Financial support and sponsorship
The study was supported by Fluid Research Fund, Christian Medical College, Vellore, Tamil Nadu, India and CMCH Institutional fund.
Conflicts of interest
There are no conflicts of interest.
| 1. |
Amagai M. Pemphigus. In: Bolognia JL, Jorizzo JL, Schaffer JV, editors. Dermatology. 3rd ed., Vol. 1. Philadelphia Elsevier Saunders; 2012. p. 461-8.
[Google Scholar]
|
| 2. |
Wojnarowska F, Venning VA. Immunobullous diseases. In: Burns T, Cox N, Griffiths C, Breathnach S, editors. Textbook of Dermatology. 8th ed., Vol. 2. West Sussex, Wiley-Blackwell; 2010. p. 40.1 - 40.62.
[Google Scholar]
|
| 3. |
Sinha AA. The genetics of pemphigus. Dermatol Clin 2011;29:381-91.
[Google Scholar]
|
| 4. |
Veldman CM, Gebhard KL, Uter W, Wassmuth R, Grötzinger J, Schultz E, et al. Tcell recognition of desmoglein 3 peptides in patients with pemphigus vulgaris and healthy individuals. J Immunol 2004;172:3883-92.
[Google Scholar]
|
| 5. |
Yan L, Wang JM, Zeng K. Association between HLA-DRB1 polymorphisms and pemphigus vulgaris: A meta-analysis. Br J Dermatol 2012;167:768-77.
[Google Scholar]
|
| 6. |
Shams S, Amirzargar AA, Yousefi M, Rezaei N, Solgi G, Khosravi F, et al. HLA class II (DRB, DQA1 and DQB1) allele and haplotype frequencies in the patients with pemphigus vulgaris. J Clin Immunol 2009;29:175-9.
[Google Scholar]
|
| 7. |
Delgado JC, Hameed A, Yunis JJ, Bhol K, Rojas AI, Rehman SB, et al. Pemphigus vulgaris autoantibody response is linked to HLA-DQB1*0503 in Pakistani patients. Hum Immunol 1997;57:110-9.
[Google Scholar]
|
| 8. |
Carcassi C, Cottoni F, Floris L, Vacca A, Mulargia M, Arras M, et al. HLA haplotypes and class II molecular alleles in Sardinian and Italian patients with pemphigus vulgaris. Tissue Antigens 1996;48:662-7.
[Google Scholar]
|
| 9. |
Lombardi ML, Mercuro O, Ruocco V, Lo Schiavo A, Lombari V, Guerrera V, et al. Common human leukocyte antigen alleles in pemphigus vulgaris and pemphigus foliaceus Italian patients. J Invest Dermatol 1999;113:107-10.
[Google Scholar]
|
| 10. |
Miyagawa S, Higashimine I, Iida T, Yamashina Y, Fukumoto T, Shirai T, et al. HLA-DRB1*04 and DRB1*14 alleles are associated with susceptibility to pemphigus among Japanese. J Invest Dermatol 1997;109:615-8.
[Google Scholar]
|
| 11. |
Saha M, Harman K, Mortimer NJ, Binda V, Black MM, Kondeatis E, et al. Pemphigus vulgaris in White Europeans is linked with HLA class II allele HLA DRB1*1454 but not DRB1*1401. J Invest Dermatol 2010;130:311-4.
[Google Scholar]
|
| 12. |
Mobini N, Yunis EJ, Alper CA, Yunis JJ, Delgado JC, Yunis DE, et al. Identical MHC markers in non-Jewish Iranian and Ashkenazi Jewish patients with pemphigus vulgaris: Possible common central Asian ancestral origin. Hum Immunol 1997;57:62-7.
[Google Scholar]
|
| 13. |
Niizeki H, Inoko H, Mizuki N, Inamoto N, Watababe K, Hashimoto T, et al. HLA-DQA1, -DQB1 and -DRB1 genotyping in Japanese pemphigus vulgaris patients by the PCR-RFLP method. Tissue Antigens 1994;44:248-51.
[Google Scholar]
|
| 14. |
Delgado JC, Yunis DE, Bozón MV, Salazar M, Deulofeut R, Turbay D, et al. MHC class II alleles and haplotypes in patients with pemphigus vulgaris from India. Tissue Antigens 1996;48:668-72.
[Google Scholar]
|
| 15. |
Daniel BS, Hertl M, Werth VP, Eming R, Murrell DF. Severity score indexes for blistering diseases. Clin Dermatol 2012;30:108-13.
[Google Scholar]
|
| 16. |
Gupta VK, Kelbel TE, Nguyen D, Melonakos KC, Murrell DF, Xie Y, et al. Aglobally available internet-based patient survey of pemphigus vulgaris: Epidemiology and disease characteristics. Dermatol Clin 2011;29:393-404.
[Google Scholar]
|
| 17. |
Kandan S, Thappa DM. Outcome of dexamethasone-cyclophosphamide pulse therapy in pemphigus: A case series. Indian J Dermatol Venereol Leprol 2009;75:373-8.
[Google Scholar]
|
| 18. |
Wilson C, Wojnarowska F, Mehra NK, Pasricha JS. Pemphigus in Oxford, UK, and New Delhi, India: A comparative study of disease characteristics and HLA antigens. Dermatology 1994;189 Suppl 1:108-10.
[Google Scholar]
|
| 19. |
Khan SW, Iftikhar N, Ahmed TA, Bashir M. HLA- DR alleles in Pakistani patients of pemphigus vulgaris. J Coll Physicians Surg Pak 2015;25:233-6.
[Google Scholar]
|
| 20. |
Wucherpfennig KW, Yu B, Bhol K, Monos DS, Argyris E, Karr RW, et al. Structural basis for major histocompatibility complex (MHC)-linked susceptibility to autoimmunity: Charged residues of a single MHC binding pocket confer selective presentation of self-peptides in pemphigus vulgaris. Proc Natl Acad Sci U S A 1995;92:11935-9.
[Google Scholar]
|
| 21. |
Lee E, Lendas KA, Chow S, Pirani Y, Gordon D, Dionisio R, et al. Disease relevant HLA class II alleles isolated by genotypic, haplotypic, and sequence analysis in North American Caucasians with pemphigus vulgaris. Hum Immunol 2006;67:125-39.
[Google Scholar]
|
| 22. |
Svecova D, Parnicka Z, Pastyrikova L, Urbancek S, Luha J, Buc M, et al. HLA DRB1* and DQB1* alleles are associated with disease severity in patients with pemphigus vulgaris. Int J Dermatol 2015;54:168-73.
[Google Scholar]
|
| 23. |
Capon F, Bharkhada J, Cochrane NE, Mortimer NJ, Setterfield JF, Reynaert S, et al. Evidence of an association between desmoglein 3 haplotypes and pemphigus vulgaris. Br J Dermatol 2006;154:67-71.
[Google Scholar]
|
| 24. |
Sarig O, Bercovici S, Zoller L, Goldberg I, Indelman M, Nahum S, et al. Population-specific association between a polymorphic variant in ST18, encoding a pro-apoptotic molecule, and pemphigus vulgaris. J Invest Dermatol 2012;132:1798-805.
[Google Scholar]
|
| 25. |
Angelini G, Bonamonte D, Lucchese A, Favia G, Serpico R, Mittelman A, et al. Preliminary data on pemphigus vulgaris treatment by a proteomics-defined peptide: A case report. J Transl Med 2006;4:43.
[Google Scholar]
|
Fulltext Views
3,466
PDF downloads
3,126





