Translate this page into:
Distribution of killer immunoglobulin-like receptor genes in HIV infected long-term non-progressors from Mumbai, India
2 Department of Microbiology, Seth G. S. Medical College and KEM Hospital, Mumbai, Maharashtra, India
Correspondence Address:
Jayanti Mania-Pramanik
National Institute for Research in Reproductive Health, J. M. Street, Parel, Mumbai - 400 012, Maharashtra
India
| How to cite this article: Chavan VR, Ansari Z, Mehta P, Mania-Pramanik J. Distribution of killer immunoglobulin-like receptor genes in HIV infected long-term non-progressors from Mumbai, India. Indian J Dermatol Venereol Leprol 2018;84:247 |
Abstract
Background: Few reports suggest the association of killer immunoglobulin-like receptors of natural killer cells with human immunodeficiency virus infection. India with world's third largest population of human immunodeficiency virus / acquired immunodeficiency syndrome, offers scope to study such association.Objective: Current study (2010-2015) was designed to evaluate if killer immunoglobulin-like receptors gene polymorphisms are associated with HIV infection outcomes specifically, with long term non progressors.
Methods: Killer immunoglobulin-like receptors genotyping was done using polymerase chain reaction - sequence-specific primer method. Viral load was measured by Cobas Taqman HIV-1 test. Estimation of CD4 counts was done using BD FACS CD4 count reagent.
Results: The activating gene frequencies identified were 3DS1 (53.8%), 2DS3 (69.2%), 2DS4 (76.9%), 2DS5 (69.2%), 2DS1 (76.9%) and 2DS2 (92.3%). The inhibitory gene frequencies were 2DL2 (92.3%), 2DL5 (76.9%), 2DL3 (69.5%), 3DL1 (84.6%), 3DL2 (92.3%) and 2DL1 (100%). The results highlight high frequency of 3DS1/3DL1 heterozygote and killer immunoglobulin-like receptor 2DS1, among these long term non progressors indicating their possible association with slow progression. Genotype analysis shows total 13 genotypes, of which 8 genotypes were identified for the first time from India. Two genotypes were unique/novel, which were unreported. All genotypes observed in this study were considered to be Bx genotype (100 %).
Limitations: A small sample size (n=13, due to a rare cohort) and the absence of control group were the limitations of this study.
Conclusions: The present study highlights the distribution of killer immunoglobulin-like receptor genes in a very rare group of human immunodeficiency virus -1 infected individuals - long term non progressors. All the long term non progressors tested show the presence of Bx haplotype and each long term non progressors has a different killer immunoglobulin-like receptor genotype.
Introduction
Natural killer cells have a great potential to kill virus infected cells without any activation or contact with antigens, with the help of their receptors known as killer immunoglobulin-like receptors.[1] Killer immunoglobulin-like receptor genes have been linked with various diseases, including human immunodeficiency virus infection.[2] HIV infected individuals may show varied progression phenotypes depending on various factors. Rapid progressors, quickly develop acquired immunodeficiency syndrome-like symptoms within 2–3 years of infection while another group of human immunodeficiency virus-infected individuals was identified as long-term non-progressors. Long-term non-progressors show no clinical or immunologic progression towards acquired immunodeficiency syndrome over an extended period, and are able to control the infection with good CD4 counts, without antiretroviral therapy. These long-term non-progressors constitute about 2.4%–5.0% of human immunodeficiency virus-infected population.[3] What prevents this group of individuals from developing acquired immunodeficiency syndrome-like features has been a matter of intense investigation. A study has reported that a combined genotype of KIR3DS1 and a subset of human leukocyte antigen-Bw4+ alleles that have isoleucine at position eighty of the major histocompatibility complex class I heavy chain (Bw4Ile80) progress more slowly to acquired immunodeficiency syndrome than those without this activating killer immunoglobulin-like receptor-human leukocyte antigen combination.[2] The present study was undertaken to evaluate the distribution of killer immunoglobulin-like receptor genes in human immunodeficiency virus-1 infected long-term nonprogressors in an attempt to highlight their possible association with the particular phenotype.
Materials and Methods
Long-term non-progressors were defined as individuals: a) who were positive for human immunodeficiency virus-1 infection for a minimum of 7 years with stable CD4 count (>600 cells/μl), b) had low levels of cell-free virus (<5000 ribonucleic acid copies/ml) in peripheral blood, c) had no history of antiretroviral therapy, and d) were asymptomatic.[4] Participants for this prospective cohort study consisted of long-term nonprogressors attending the Integrated Counseling and Testing Centre (Shakti Clinic) of the Seth Gordhandas Sunderdas Medical College and King Edward Memorial Hospital, Mumbai, between January 2010 and December 2015 for regular human immunodeficiency virus testing, CD4 count and disease management. A written informed consent was obtained from all the enrolled study participants. CD4 count estimation was done using BD FACS Calibur Flow cytometer. Viral load analysis in blood samples was done by isolating total viral nucleic acid using the MagNA Pure Compact Nucleic Acid Automated System (Roche Diagnostic, Mannheim, Germany). Subsequently, the viral load was measured by COBAS TaqMan human immunodeficiency virus-1 test, version 2.0 real-time polymerase chain reaction (Roche Molecular Systems, Branchburg, NJ, USA). The detection limit of the assay was up to 34 copies of ribonucleic acid/ml of plasma. To assess the quality of the viral load results, randomly selected samples (n = 6), such as those with undetectable values, high viral load values and intermediate values were reassessed.
Isolated deoxyribonucleic acid from each blood sample was used for killer immunoglobulin-like receptor genotyping using polymerase chain reaction with sequence-specific primer method (Inno-Train, Diagnostic GmbH, Germany), to detect presence or absence of 16 killer immunoglobulin-like receptor genes, namely, 2DL1, 2DL2, 2DL3, 2DL4, 2DL5, 2DS1, 2DS2, 2DS3, 2DS4, 2DS5, 3DL1, 3DL2, 3DL3, 3DS1 and two pseudogenes: 2DP1 and 3DP1. In case of unique genotypes, genotyping was repeated twice. Killer immunoglobulin-like receptor haplotypes were identified using the website of European Bioinformatics Institute (http://www.ebi.ac.uk/ipd/kir). Group A haplotypes were identified by the presence of one or more of the following genes: KIR2DL1, KIR2DL3, KIR3DL1 and KIR2DS4 and the absence of group B haplotype genes such as KIR2DL2, KIR2DL5, KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS5 and KIR3DS1. Killer immunoglobulin-like receptor genotypes consist of two killer immunoglobulin-like receptor haplotypes. AA genotypes were identified by the absence of all group B haplotype genes. If any one B haplotype gene was present, then they were characterized as having AB and BB genotypes. However, AB and BB genotypes were very difficult to distinguish from each other and therefore collectively annotated as Bx. The genotype reference of each individual (genotype identification numbers) was searched using a freely available interactive online database at the website (http://www.allelefrequencies.net:).[5] Individual gene frequencies of killer immunoglobulin-like receptor genes were calculated by direct counting. Average, percentage, median and standard errors were calculated using descriptive statistics.
Results
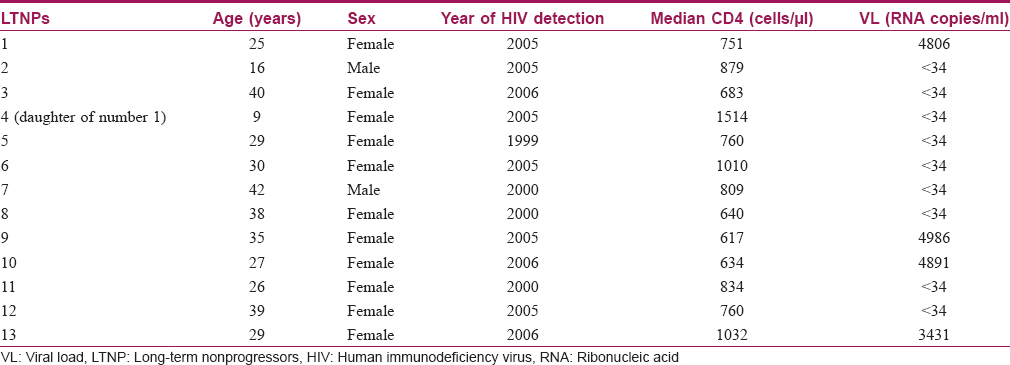
During the study period, 13 long-term non-progressors (2 males, 11 females) were enrolled as per defined criteria. All the long-term non-progressors (mean average age: 29.6 ± 9.6 years) had a controlled viral load over a period of time. The median CD4 count in long-term non-progressors was 840 ± 241 cells/μl (range: 634–1514 cells/μl). None of them had any opportunistic or co-infections. Detailed characteristics of these long-term nonprogressors are presented in [Table - 1].
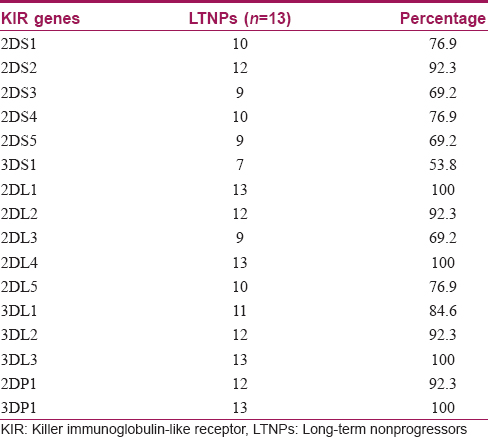
The killer immunoglobulin-like receptor genes distribution in long-term nonprogressors is presented in [Table - 2]. The activating gene frequencies were 3DS1 (53.8%), 2DS3 (69.2%), 2DS4 (76.9%), 2DS5 (69.2%), 2DS1 (76.9%) and 2DS2 (92.3%). The inhibitory gene frequencies were 2DL2 (92.3%), 2DL5 (76.9%), 2DL3 (69.2%), 3DL1 (84.6%) and 2DL1 (100%). The frequency of 3DS1/3DL1 heterozygote was high (53.9%). Frequencies of inhibitory genes (62.0%) were significantly (P = 0.001) higher than that of activating genes (38%). The framework genes 2DL4 and 3DL3 were present in all long-term non-progressors. Genotype identification analysis identified 13 genotypes (genotype identification numbers) in total, of which 8 genotype identification numbers were identified for the first time from India, along with 2 unique/novel genotypes [Figure - 1]. All the genotypes observed in this study were considered to be Bx genotype (100%). All the killer immunoglobulin-like receptor genotypes identified in long-term non-progressors had a similar carrier frequency of 7.7%. The phenotypic frequency in all long-term non-progressors varied from 53.8% to 100%.
 |
| Figure 1: Killer immunoglobulin-like receptor genotypes in long-term nonprogressor |
Discussion
Although several host, viral and environmental factors influence the infection outcome, in the present study, we have focused on killer immunoglobulin-like receptor genes and their association with human immunodeficiency virus pathogenesis in a cohort of long-term non-progressors.
It has been reported that viral load is the strongest predictor of human immunodeficiency virus disease progression.[6] In this study, nine long-term non-progressors enrolled (genotype identification number: 13, 93, 63, 120, 232, 325, 381, 463 and 1 unique genotype) had <34 copies/ml and 4 long-term nonprogressors had <5000 copies/ml of plasma viral load.
Our analysis highlights a high frequency of 3DS1/L1 heterozygotes among long-term non-progressors. A similar observation was made in a study on long-term non-progressors from China, where KIR3DS1/L1 heterozygote condition was associated with slower CD4 cell decline, indicating its possible protective effect.[7] This high frequency of 3DS1/L1 was also observed among the exposed seronegative spouses, whereas KIR2DS1 was significantly associated with low viral load in human immunodeficiency virus seropositive patients in a discordant couple cohort from India.[8] In the present study too, high percentage of the long-term non-progressors (76.9%) showed the presence of KIR2DS1 gene, indicating its possible association in viral load control. Earlier a study in South African population had also shown the association of KIR2DS1 gene with decreasing human immunodeficiency virus-1 viral load.[9]
In our long-term nonprogressors cohort, none had opportunistic/coinfections similar to the observation among long-term non-progressors from South India. Their median CD4 count was also comparable to the human immunodeficiency virus-1 infected population from Southern India.[10],[11]
Till date, 442 distinct killer immunoglobulin-like receptor genotypes have been identified from 125 populations, of which only 10 belong to AA genotype. The current study showed Bx genotype in this long-term non-progressors cohort, as was observed in previous Indian studies.[12]
Another interesting finding associated with this study was the predominance of females in the long-term non-progressors group, similar to a report on the natural history of human immunodeficiency virus disease, which showed that not only is the number of human immunodeficiency virus-infected males more, but their survival rate is also significantly low compared to human immunodeficiency virus positive females.[11]
The limitations of the study were the small number of long-term non-progressors and the absence of a control group. Since long-term non-progressors normally constitute only 2.4%–5.0% of the human immunodeficiency virus-infected population, the sample size included was small.[3] Each long-term non-progressor showed a distinct killer immunoglobulin-like receptor genotype suggesting that it might have a role in human immunodeficiency virus pathogenesis and disease progression. This knowledge could contribute to an increased understanding of the role of genetic factors in human immunodeficiency virus-1 infection.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
| 1. |
Parham P. Taking license with natural killer cell maturation and repertoire development. Immunol Rev 2006;214:155-60.
[Google Scholar]
|
| 2. |
Martin MP, Gao X, Lee JH, Nelson GW, Detels R, Goedert JJ, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet 2002;31:429-34.
[Google Scholar]
|
| 3. |
Okulicz JF, Marconi VC, Landrum ML, Wegner S, Weintrob A, Ganesan A, et al. Clinical outcomes of elite controllers, viremic controllers, and long-term nonprogressors in the US Department of Defense HIV natural history study. J Infect Dis 2009;200:1714-23.
[Google Scholar]
|
| 4. |
Sheppard HW, Lang W, Ascher MS, Vittinghoff E, Winkelstein W. The characterization of non-progressors: Long-term HIV-1 infection with stable CD4+T-cell levels. AIDS 1993;7:1159-66.
[Google Scholar]
|
| 5. |
Middleton D, Diler AS, Meenagh A, Sleator C, Gourraud PA. Killer immunoglobulin-like receptors (KIR2DL2 and/or KIR2DS2) in presence of their ligand (HLA-C1 group) protect against chronic myeloid leukaemia. Tissue Antigens 2009;73:553-60.
[Google Scholar]
|
| 6. |
Arnaout RA, Lloyd AL, O'Brien TR, Goedert JJ, Leonard JM, Nowak MA. A simple relationship between viral load and survival time in HIV-1 infection. Proc Natl Acad Sci U S A 1999;96:11549-53.
[Google Scholar]
|
| 7. |
Jiang Y, Chen O, Cui C, Zhao B, Han X, Zhang Z, et al. KIR3DS1/L1 and HLA-Bw4-80I are associated with HIV disease progression among HIV typical progressors and long-term nonprogressors. BMC Infect Dis 2013;13:405.
[Google Scholar]
|
| 8. |
Chavan VR, Chaudhari D, Ahir S, Ansari Z, Mehta P, Mania-Pramanik J. Variations in KIR genes: A study in HIV-1 serodiscordant couples. Biomed Res Int 2014;2014:891402.
[Google Scholar]
|
| 9. |
Wong AH, Williams K, Reddy S, Wilson D, Giddy J, Alter G, et al. Alterations in natural killer cell receptor profiles during HIV type 1 disease progression among chronically infected South African adults. AIDS Res Hum Retroviruses 2010;26:459-69.
[Google Scholar]
|
| 10. |
Shanmugasundaram U, Murugavel KG, Shankar EM, Balakrishnan P, Solomon S, Kumarasamy N. Laboratory characteristics of HIV-1 clade C-infected long-term non-progressors at a tertiary human immunodeficiency virus care centre in South India. J Med Microbiol 2008;57:913-5.
[Google Scholar]
|
| 11. |
Kumarasamy N, Solomon S, Flanigan TP, Hemalatha R, Thyagarajan SP, Mayer KH. Natural history of human immunodeficiency virus disease in Southern India. Clin Infect Dis 2003;36:79-85.
[Google Scholar]
|
| 12. |
Rajalingam R, Du Z, Meenagh A, Luo L, Kavitha VJ, Pavithra-Arulvani R, et al. Distinct diversity of KIR genes in three Southern Indian populations: Comparison with world populations revealed a link between KIR gene content and pre-historic human migrations. Immunogenetics 2008;60:207-17.
[Google Scholar]
|
Fulltext Views
1,928
PDF downloads
2,335





