Translate this page into:
Genetic and molecular aspects of androgenetic alopecia
2 Universidad Autónoma de Nuevo León, Facultad de Medicina, Departamento de Dermatología, Monterrey, Mexico
3 Tecnológico de Monterrey, Escuela de Medicina y Ciencias de la Salud, Monterrey, Mexico
Correspondence Address:
Augusto Rojas-Martínez
Tecnológico de Monterrey, Escuela de Medicina y Ciencias de la Salud, 3er. Piso CITES, Ave. I. Morones Prieto 3000 Poniente, Col. Los Doctores, Monterrey, 64710
Mexico
| How to cite this article: Martinez-Jacobo L, Villarreal-Villarreal CD, Ortiz-López R, Ocampo-Candiani J, Rojas-Martínez A. Genetic and molecular aspects of androgenetic alopecia. Indian J Dermatol Venereol Leprol 2018;84:263-268 |
Abstract
Androgenetic alopecia is the most common form of progressive hair loss in humans. A genetic predisposition and hormonal status are considered as major risk factors for this condition. Several recent advances in molecular biology and genetics have increased our understanding of the mechanisms of hair loss in androgenetic alopecia. We review these advances and examine the trends in the genetic and molecular aspects of androgenetic alopecia.
Introduction
Androgenetic alopecia is the most common form of hair loss in humans affecting 80% of Caucasian men and 50% of Caucasian women.[1] Hair loss typically begins with bitemporal recession of the frontal hairline, followed by diffuse hair thinning at the vertex, and eventual complete loss of hair at the center of the vertex. The bald patch at the vertex subsequently joins the frontal receding hairline, leaving an island of hair on the frontal scalp. This island finally disappears leaving hair only in the parietal and occipital zones. Other less common patterns include more rapid hair loss over the vertex than the frontal area, frontal hairline loss before the vertex bald patch develops and also, a Ludwig-type pattern with preservation of the frontal hair line.[2] The Hamilton–Norwood scale is used to assess the extent and severity of androgenetic alopecia in men,[3] whereas the Ludwig scale is preferred for women.
Both men and women have higher levels of androgen receptors and alpha-reductase type I and II activities in the frontal area of the scalp as compared to hair follicles located in the occipital area which have higher aromatase levels. The alpha-reductase type I and II activity in frontal hair follicles is three times greater in men than in women.[4] Thus, male androgenetic alopecia is considered an androgen dependent condition, but the role of androgen signaling in women remains uncertain.[5]
Efforts have been made to elucidate the molecular mechanisms of androgenetic alopecia. Different expression profiles have been proposed in the areas affected by androgenetic alopecia, and various loci have been shown to be associated with this condition, suggesting that nonandrogen-dependent pathways may be involved in the pathophysiology of androgenetic alopecia.
This review focuses on the recent trends in the molecular and genetic aspects involved in the pathogenesis of androgenetic alopecia.
Etiology
Each hair originates in a hair follicle, and a cyclic process known as the hair growth cycle defines its individual and asynchronous growth. This cycle consists of four phases: (1) the anagen or growth phase, which is the longest and lasts 2 to 7 years, (2) the catagen or transition phase, which lasts approximately 2 weeks and includes hair follicle involution due to apoptosis, (3) the telogen or resting phase, which lasts 12 weeks when old hair is removed and (4) the exogen phase, which is the release phase of the telogen hair.[12] Miniaturization of the hair follicle is the hallmark of androgenetic alopecia.[13] It occurs at some point between the late catagen or early anagen phase, affecting the dermal papilla and the dermal sheath, resulting in a smaller follicle and a reduced anagen phase.[14] This anomaly is usually irreversible, although partial regrowth and some reversal of miniaturization is possible in some instances.
The different processes involved in the pathogenesis of androgenetic alopecia are shown in [Figure - 1]. The condition is clearly associated with the activity of androgenic hormones and genetic predisposition. It demonstrates a pattern of familial aggregation, but can occur in several individuals within a family without monogenic inheritance, and is therefore considered a complex phenotype. Twin studies have demonstrated a heritability of 0.81, suggesting that genetics plays an important role in its presentation and progression.[6]
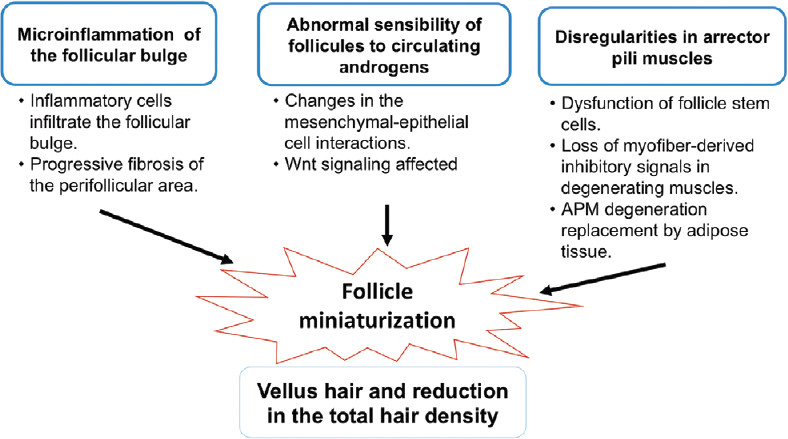 |
| Figure 1: Different factors involved in the pathogenesis of androgenetic alopecia. Microinflammation, abnormal sensitivity to androgens and irregularities in the arrector pilli muscles leads to miniaturization of hair follicles, clinically defined as vellus hair |
It has been assumed that androgenetic alopecia is the result of the abnormal sensitivity of hair follicles of the scalp to circulating androgens,[7] owing to an increase in the number of androgen receptors.[2] The enzyme 5-alpha reductase has two isoforms, types 1 and 2, which catalyze the conversion of testosterone to 5-alpha–dihydrotestosterone. It is believed that both isoforms play a role in the metabolism and action of androgen, and their expression varies depending on the site of the body. Liu and Yamauchi [8] found a higher expression of 5-alpha reductase type 1 in hair follicles, suggesting that they play a key role in androgen-regulated hair growth.
Androgens alter mesenchyme-epithelial cell interactions within the follicle, thus affecting hair growth, dermal papilla size, dermal papilla cells, and keratinocyte and melanocyte activities.[9] The Wnt signaling pathway regulates cells in the dermal papilla and may play a pivotal role in the action of androgen on hair growth. However, the underlying molecular mechanisms of androgen-related actions remain largely unknown.
The replacement of terminal hair by vellus hair (vellus hair is defined as hair <30 μm in diameter and <30 μm in length; transitional hair width between 30 and 40 μm; terminal hair >40 μm) and a reduction in the total hair density (hair/square centimeter) are the major clinical features of androgenetic alopecia.[10] The replacement of terminal follicles by vellus follicles is seen histologically too, along with a perifollicular infiltrate of macrophages, an increase in the size of sebaceous glands and dermal thinning.[11]
Whether follicle miniaturization occurs mainly due to the activity of androgens is still a matter of debate. Whiting showed that miniaturization can be reversed in a single hair cycle in patients treated with finasteride, supporting the involvement of androgens in androgenetic alopecia. It is hypothesized that the miniaturization seen in patterned hair loss may be the direct result of reduction in the cell number and, hence, in the size of the dermal papilla.[14] Yazdabadi et al. suggested that the arrector pili muscle serves as a source of stem cells to maintain the follicle, stimulating stem cell populations in the bulge or dermal sheath. The loss of contact in miniaturized follicles between the arrector pili muscle and the bulge produces miniaturization by disrupting the function of stem cells residing in the follicle.[15]
The stem cells of the hair follicle are located in the bulge where they periodically alternate between activated and quiescent phases to maintain the stem cell population and produce new hair follicles. Wang et al. noted that when murine hair follicle stem cells were active, the FOXC1 gene was highly expressed, maintaining stem cell adhesion and promoting transition to the quiescent state. FOXC1 is a transcription factor involved in the regulation of embryonic development and the eye. It also activates the signaling of Nfatc1 and bone morphogenetic proteins, which play a role in the maintenance and development of hair follicles.[16]
The arrector pili muscle plays an important role in maintaining follicle integrity by holding together each of the hair follicles in a follicular unit at the isthmus level.[17] Torkamani et al. found that the arrector pili muscle degenerates and is replaced by adipose tissue in androgenetic alopecia.[18] It is not clear how arrector pili muscle degeneration and fat infiltration are mechanistically related to follicle miniaturization and hair loss. It has been speculated that adipocytes derived from the aberrant differentiation of the remaining progenitor cells in the arrector pili muscle can cause follicle miniaturization.[19] Mesenchymal progenitor cells in muscle tissue with ectopic fat deposition suggest that the loss of myofiber-derived inhibitory signals in degenerating muscles permits cellular adipose differentiation.[20] Rushton et al. have proposed the existence of growth restricted (dormant/kenogen) nonvellus hair follicles that are re-activated by medical treatments in FPHL and MPHL.[21] All these findings support the role of follicle miniaturization in the pathogenesis of androgenetic alopecia. Further studies are needed to define the role of follicle miniaturization in androgenetic alopecia more completely.
The role of microinflammation in the pathogenesis of androgenetic alopecia has been investigated by Jaworsky et al. who showed that the inflammatory cell infiltrate in the follicular bulge produces a progressive fibrosis of the perifollicular zone resulting in injury to follicular stem cells, impairment of normal hair cycling and finally, hair loss.[22]
Genetics and Androgenetic Alopecia
Although androgenetic alopecia is mediated by androgens, genetic predisposition plays an important role in its etiology. The genetics of androgenetic alopecia is complex. The AR and 5-alpha reductase genes are attractive candidates for androgenetic alopecia.
Several studies have focused on genes related to the sex-steroid pathways guided by reports of differential levels of sex-steroid receptor expression and metabolizing enzymes between balding and occipital regions of the scalp. There is evidence that both 5-alpha reductase enzymes and the androgen receptor are highly expressed in balding follicles as compared with nonbalding follicles on the same scalp [23] which is due to the different genes that encode 5-alpha reductase type I and II and androgen receptors being expressed in these locations.[24]
Recent genome-wide association studies in AGA have identified strong association signals in the X chromosome. Both the AR gene and the ectodysplasin A2 receptor (AR/EDA2R locus in Xq11-q12) showed strong signals for AGA.[25]
It has been estimated that the AR gene may confer up to 40% of the total genetic risk, which is considered a high level of risk for a single gene.[26] There have been several efforts to determine whether the AR gene is associated with male pattern baldness. Single nucleotide polymorphisms, copy number variations and triplet repeats are among the polymorphisms that have been studied in relation to AGA. Ellis et al. compared the allelic frequencies between two polymorphisms in exon 1 of the AR gene, the StuI restriction site and CAG and GGC triplet repetition polymorphisms. This study found that the Stu restriction polymorphism was present in 98.1% of younger and 92.3% of older men with androgenetic alopecia as compared to 76.6% of non-bald controls. The combination of fewer trinucleotide repeats was more common in men with baldness (P = 0.03) suggesting that these markers are a close functional variant involved in the polygenic determination of male pattern baldness.[24] In 2005, Hillmer et al. replicated this work and showed that the leading candidate for AGA was the polyglycine GGN triplet repeat.[27] However, Ellis et al. found that these polyglycine repeats do not confer susceptibility to AGA [28] and analyses of copy number variations in AR also suggest that polyglycine repeats are not implicated in AGA .
Prodi et al. showed that the AR and EDA2R genes on the X chromosome were strongly associated with AGA. SNP rs1385699, which is located in EDA2R, displayed the best association signal (P = 3.9 × 10-19), while the variant located in AR rs6152 showed lower significance (P = 4.17 × 10-12). Although the role of EDA2R in androgenetic alopecia is not clear, statistical analyses show that the association of markers in EDA2R and AR appear to be the result of linkage disequilibrium.[29] The location of AR on the X chromosome and the strong association signal of EDA2R highlight the importance of the maternal lineage in androgenetic alopecia inheritance.[27]
These findings emphasize the importance of the androgen receptor gene responsible for the increased risk of androgenetic alopecia in males which have been confirmed in multiple independent studies.[27] However, a gene effect has not been demonstrated in women.
Genome-Wide Association Studies and Risk Loci for Androgenetic Alopecia
Genome-wide association studies and meta-analysis have been used to evaluate the complex inheritance of androgenetic alopecia. Genome-wide association studies involve scanning markers across a complete set of genomic DNA of many cases and controls to determine genetic variations associated with a particular trait or disease. These studies use microarray technology to identify genetic markers or candidate genes.[30] Meta-analyses are a powerful tools used to combine the results of genome-wide association studies from studies with similar designs across different populations to demonstrate a more significant association.[31]
Heilmann et al. suggested a polygenic component to androgenetic alopecia that may be part of the complex biological pathways associated with androgenetic alopecia.[1] They identified four risk loci for androgenetic alopecia located in 2q35, 3q25.1, 5q33.3 and 12p12.1. The strongest association signal was observed in 2q35 (P = 3.33 × 10-15). This locus contains the WNT10A gene, which is expressed in the bulge during the anagen phase of the hair growth cycle and has been shown to have a genotypic effect on hair follicle expression.[25]
A meta-analysis by Li et al. identified 6 new risk loci for androgenetic alopecia in 1p36.22, 2q37, 7p21.1, 7q11.22, 17q21.31 and 18q21.1 and a strong association for androgenetic alopecia in 20p11 and the AR gene (rs2497938: OR = 2.20, P = 2.40 × 10-9).[32] The risk locus for androgenetic alopecia at 20p11 had earlier been identified by Liang et al. in a Chinese population.[33] A risk locus at 3q26 was identified in a German population [34] and a polymorphism in the APCDD1 gene located in 18p11.2 has been also been associated with androgenetic alopecia (rs3185480). This latter polymorphism is interesting because APCDD1 is a Wnt signaling inhibitor.[35] The identification of a new susceptibility autosomal gene in these autosomal loci suggests that nonandrogen-dependent pathways are also involved in androgenetic alopecia pathogenesis.[36]
Pathways Signaling Related to Androgenetic Alopecia
Relatively few studies have used expression analyses because of difficulties in obtaining scalp biopsies. Various genes such as BMP2, ephrinA3, PGDS, PGD2, BDNF, neurotrophin-3 protein, neural growth factor-β, ASS1 and GSN have been found to be differentially expressed in dermal papilla cell culture and scalp biopsy samples from patients with androgenetic alopecia. These genes may be involved in the development of androgenetic alopecia as either hair growth promoters or inhibitors, although more studies are needed to confirm these results.[37],[38]
Mirmirani et al. identified 38 differentially expressed genes between the scalp vertex and frontal regions of 16 patients with androgenetic alopecia. Among the overexpressed genes were MSL3L2, CD209, MUC7, SLC6A14 and ANKRD20B. The subexpressed genes included DUSP1, FOS, FOSB, CYR61, HBB, EGR1, ZFP36, MS4A1, IGLI3, ATF3, PSG3, EFCAB4B, KRTAP as well as various noncoding RNAs. These authors suggest a specific expression signature for these two regions.[39]
Garza et al. conducted a microarray global expression analysis with biopsies from the balding and occipital regions of 5 patients with androgenetic alopecia and found 250 differentially expressed transcripts. However, only the overexpression of prostaglandin synthase (PGDS) was relevant to the region of baldness and concluded that the PGDS product prostaglandin D2 (PGD2) inhibits hair growth by inducing a premature catagen phase.[37]
Heilmann et al. however, felt that there is no genetic evidence for the contribution of prostaglandins to the etiology of androgenetic alopecia as genome-wide association studies have not shown any association signals near these genes (PGDS and PGD2) and suggested the role of the Wnt pathway because of the genotypic effect of WNT10A expression.[25] The involvement of Wnt was also proposed by Leirós et al. who found that androgen action inhibits the canonical Wnt/B- catenin, leading to follicle miniaturization [Figure - 2]a,[Figure - 2]b,[Figure - 2]c.[40]
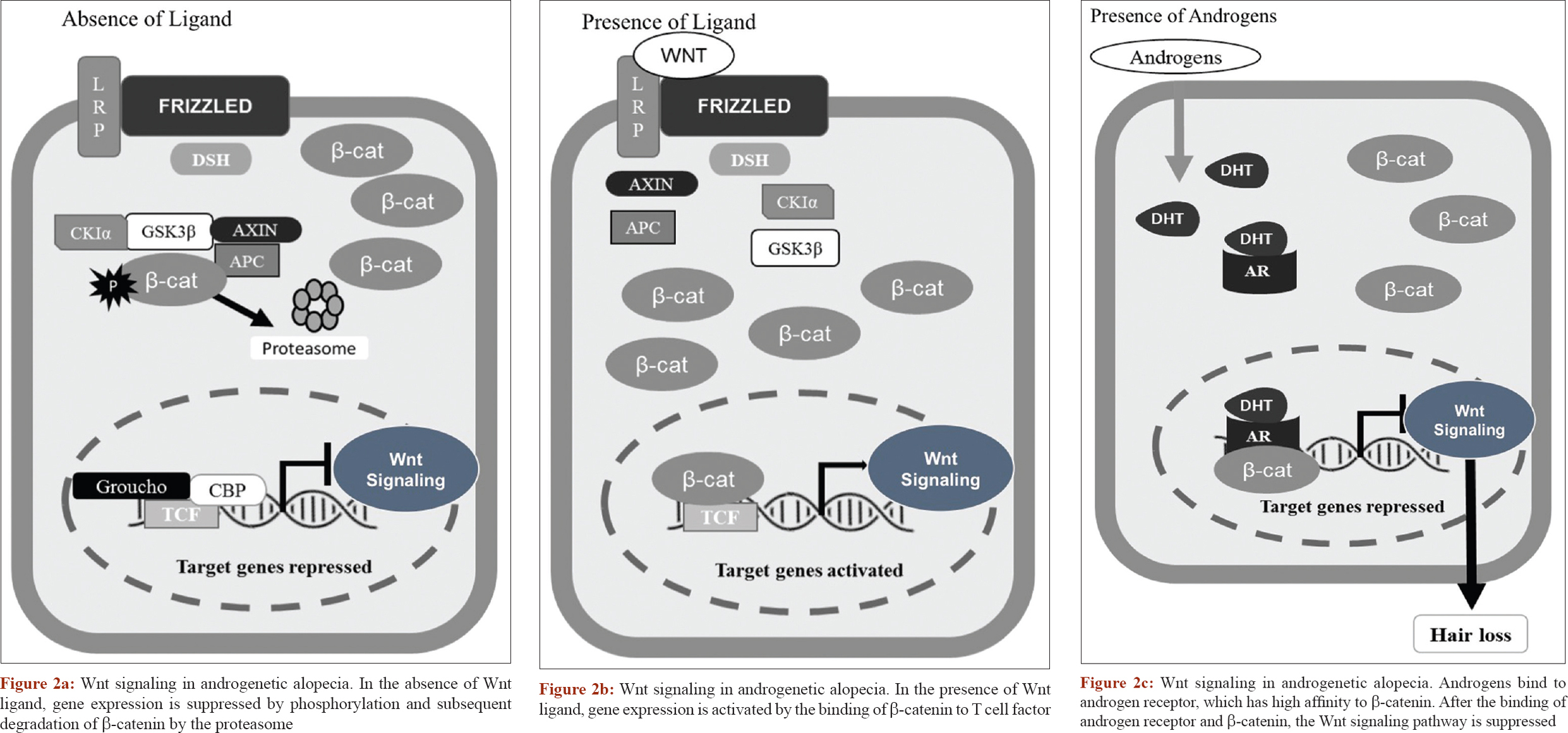 |
| Figure 2: |
The Notch signaling pathway is also involved in androgenetic alopecia. Midorikawa et al. found that increased androgen levels resulted in a negative feedback of gene expression of the Notch pathway, leading to miniaturization of the hair follicle and overexpression of the AR gene.[38] Both the Notch and the Wnt pathways are directly affected by androgen expression in androgenetic alopecia.
Epigenetic Changes in Androgenetic Alopecia
It has been demonstrated that epigenetic mechanisms involved in histone or DNA methylation modulate the accessibility of genes to the transcriptional machinery and are involved in gene regulation activities in which genomic DNA sequences remain unchanged.[41] However, few studies have evaluated the role of epigenetics on hair follicle physiology and androgenetic alopecia etiology.
Recent studies have focused on methylation patterns of specific genes such as the AR gene. Cobb et al. investigated methylation patterns on the AR gene in occipital hair follicles and affected follicles [42] and observed an increase in AR gene methylation in occipital follicles, suggesting that increased AR gene methylation protects the occipital follicles against miniaturization and hair loss. Another study demonstrated that mice lacking the expression of DNA methyl-transferase 1 (DNMT1) in the skin produced a baldness phenotype.[30]DNMT1 is involved in the establishment and regulation of methylation patterns for tissue-specific cytokines, suggesting that the activity of this gene may be relevant for the development of androgenetic alopecia.
Conclusion
Androgenetic alopecia is a multifactorial dermatological condition with a complex genetic inheritance. Miniaturization of the hair follicles is the hallmark of androgenetic alopecia. Microinflammation in the follicular bulge plays an important role in the disruption of stem cells resulting in fibrosis of the perifollicular zone and irreversible miniaturization.
Maternal inheritance in androgenetic alopecia may largely be explained by the locus AR in the X chromosome.
Androgens are crucial in androgenetic alopecia as they inhibit the expression of Wnt/B-catenin and produce a negative feedback in Notch signaling, both leading to miniaturization of the hair follicle. The discovery of overexpression of the prostaglandin synthase gene (PGDS) and overproduction of prostaglandin D2 (PGD2) is being explored for future potential therapies.
More studies of androgenetic alopecia at the molecular level are necessary as studies performed to analyze epigenetics are gene-specific. It will be relevant to know the global methylation profile of patients with androgenetic alopecia to identify other genes involved in this condition. Despite these important recent contributions, there is still much to learn about androgenetic alopecia molecular pathophysiology.
Acknowledgement
We thank Dr. Sergio Lozano for the review of this article.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
| 1. |
Heilmann S, Brockschmidt FF, Hillmer AM, Hanneken S, Eigelshoven S, Ludwig KU, et al. Evidence for a polygenic contribution to androgenetic alopecia. Br J Dermatol 2013;169:927-30.
[Google Scholar]
|
| 2. |
Sinclair R. Male pattern androgenetic alopecia. BMJ 1998;317:865-9.
[Google Scholar]
|
| 3. |
Guarrera M, Cardo P, Arrigo P, Rebora A. Reliability of hamilton-norwood classification. Int J Trichology 2009;1:120-2.
[Google Scholar]
|
| 4. |
Sawaya ME, Price VH. Different levels of 5alpha-reductase type I and II, aromatase, and androgen receptor in hair follicles of women and men with androgenetic alopecia. J Invest Dermatol 1997;109:296-300.
[Google Scholar]
|
| 5. |
Ioannides D, Lazaridou E. Female pattern hair loss. Curr Probl Dermatol 2015;47:45-54.
[Google Scholar]
|
| 6. |
Nyholt DR, Gillespie NA, Heath AC, Martin NG. Genetic basis of male pattern baldness. J Invest Dermatol 2003;121:1561-4.
[Google Scholar]
|
| 7. |
Kaufman KD. Androgens and alopecia. Mol Cell Endocrinol 2002;198:89-95.
[Google Scholar]
|
| 8. |
Liu S, Yamauchi H. Different patterns of 5alpha-reductase expression, cellular distribution, and testosterone metabolism in human follicular dermal papilla cells. Biochem Biophys Res Commun 2008;368:858-64.
[Google Scholar]
|
| 9. |
Randall VA. Androgens and hair growth. Dermatol Ther 2008;21:314-28.
[Google Scholar]
|
| 10. |
Rushton H, James KC, Mortimer CH. The unit area trichogram in the assessment of androgen-dependent alopecia. Br J Dermatol 1983;109:429-37.
[Google Scholar]
|
| 11. |
Lattanand A, Johnson WC. Male pattern alopecia a histopathologic and histochemical study. J Cutan Pathol 1975;2:58-70.
[Google Scholar]
|
| 12. |
Van Neste D, Leroy T, Conil S. Exogen hair characterization in human scalp. Skin Res Technol 2007;13:436-43.
[Google Scholar]
|
| 13. |
Jahoda CA. Cellular and developmental aspects of androgenetic alopecia. Exp Dermatol 1998;7:235-48.
[Google Scholar]
|
| 14. |
Whiting DA. Possible mechanisms of miniaturization during androgenetic alopecia or pattern hair loss. J Am Acad Dermatol 2001;45:S81-6.
[Google Scholar]
|
| 15. |
Yazdabadi A, Whiting D, Rufaut N, Sinclair R. Miniaturized hairs maintain contact with the arrector pili muscle in alopecia areata but not in androgenetic alopecia: A model for reversible miniaturization and potential for hair regrowth. Int J Trichology 2012;4:154-7.
[Google Scholar]
|
| 16. |
Wang L, Siegenthaler JA, Dowell RD, Yi R. Foxc1 reinforces quiescence in self-renewing hair follicle stem cells. Science 2016;351:613-7.
[Google Scholar]
|
| 17. |
Poblet E, Jiménez F, Ortega F. The contribution of the arrector pili muscle and sebaceous glands to the follicular unit structure. J Am Acad Dermatol 2004;51:217-22.
[Google Scholar]
|
| 18. |
Torkamani N, Rufaut NW, Jones L, Sinclair R. Destruction of the arrector pili muscle and fat infiltration in androgenic alopecia. Br J Dermatol 2014;170:1291-8.
[Google Scholar]
|
| 19. |
Brack AS, Conboy MJ, Roy S, Lee M, Kuo CJ, Keller C, et al. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science 2007;317:807-10.
[Google Scholar]
|
| 20. |
Uezumi A, Fukada S, Yamamoto N, Takeda S, Tsuchida K. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat Cell Biol 2010;12:143-52.
[Google Scholar]
|
| 21. |
Hugh Rushton D, Norris MJ, Van Neste D. Hair regrowth in male and female pattern hair loss does not involve the conversion of vellus hair to terminal hair. Exp Dermatol 2016;25:482-4.
[Google Scholar]
|
| 22. |
Jaworsky C, Kligman AM, Murphy GF. Characterization of inflammatory infiltrates in male pattern alopecia: Implications for pathogenesis. Br J Dermatol 1992;127:239-46.
[Google Scholar]
|
| 23. |
Hibberts NA, Howell AE, Randall VA. Balding hair follicle dermal papilla cells contain higher levels of androgen receptors than those from non-balding scalp. J Endocrinol 1998;156:59-65.
[Google Scholar]
|
| 24. |
Ellis JA, Stebbing M, Harrap SB. Polymorphism of the androgen receptor gene is associated with male pattern baldness. J Invest Dermatol 2001;116:452-5.
[Google Scholar]
|
| 25. |
Heilmann S, Kiefer AK, Fricker N, Drichel D, Hillmer AM, Herold C, et al. Androgenetic alopecia: Identification of four genetic risk loci and evidence for the contribution of WNT signaling to its etiology. J Invest Dermatol 2013;133:1489-96.
[Google Scholar]
|
| 26. |
Price VH. Treatment of hair loss. N Engl J Med 1999;341:964-73.
[Google Scholar]
|
| 27. |
Hillmer AM, Hanneken S, Ritzmann S, Becker T, Freudenberg J, Brockschmidt FF, et al. Genetic variation in the human androgen receptor gene is the major determinant of common early-onset androgenetic alopecia. Am J Hum Genet 2005;77:140-8.
[Google Scholar]
|
| 28. |
Ellis JA, Scurrah KJ, Cobb JE, Zaloumis SG, Duncan AE, Harrap SB, et al. Baldness and the androgen receptor: The AR polyglycine repeat polymorphism does not confer susceptibility to androgenetic alopecia. Hum Genet 2007;121:451-7.
[Google Scholar]
|
| 29. |
Prodi DA, Pirastu N, Maninchedda G, Sassu A, Picciau A, Palmas MA, et al. EDA2R is associated with androgenetic alopecia. J Invest Dermatol 2008;128:2268-70.
[Google Scholar]
|
| 30. |
Li J, Jiang TX, Hughes MW, Wu P, Yu J, Widelitz RB, et al. Progressive alopecia reveals decreasing stem cell activation probability during aging of mice with epidermal deletion of DNA methyltransferase 1. J Invest Dermatol 2012;132:2681-90.
[Google Scholar]
|
| 31. |
Russo MW. How to review a meta-analysis. Gastroenterol Hepatol (N Y) 2007;3:637-42.
[Google Scholar]
|
| 32. |
Li R, Brockschmidt FF, Kiefer AK, Stefansson H, Nyholt DR, Song K, et al. Six novel susceptibility loci for early-onset androgenetic alopecia and their unexpected association with common diseases. PLoS Genet 2012;8:e1002746.
[Google Scholar]
|
| 33. |
Liang B, Yang C, Zuo X, Li Y, Ding Y, Sheng Y, et al. Genetic variants at 20p11 confer risk to androgenetic alopecia in the Chinese Han population. PLoS One 2013;8:e71771.
[Google Scholar]
|
| 34. |
Hillmer AM, Flaquer A, Hanneken S, Eigelshoven S, Kortüm AK, Brockschmidt FF, et al. Genome-wide scan and fine-mapping linkage study of androgenetic alopecia reveals a locus on chromosome 3q26. Am J Hum Genet 2008;82:737-43.
[Google Scholar]
|
| 35. |
Shimomura Y, Agalliu D, Vonica A, Luria V, Wajid M, Baumer A, et al. APCDD1 is a novel wnt inhibitor mutated in hereditary hypotrichosis simplex. Nature 2010;464:1043-7.
[Google Scholar]
|
| 36. |
Rathnayake D, Sinclair R. Male androgenetic alopecia. Expert Opin Pharmacother 2010;11:1295-304.
[Google Scholar]
|
| 37. |
Garza LA, Liu Y, Yang Z, Alagesan B, Lawson JA, Norberg SM, et al. Prostaglandin D2 inhibits hair growth and is elevated in bald scalp of men with androgenetic alopecia. Sci Transl Med 2012;4:126ra34.
[Google Scholar]
|
| 38. |
Midorikawa T, Chikazawa T, Yoshino T, Takada K, Arase S. Different gene expression profile observed in dermal papilla cells related to androgenic alopecia by DNA macroarray analysis. J Dermatol Sci 2004;36:25-32.
[Google Scholar]
|
| 39. |
Mirmirani P, Consolo M, Oyetakin-White P, Baron E, Leahy P, Karnik P, et al. Similar response patterns to topical minoxidil foam 5% in frontal and vertex scalp of men with androgenetic alopecia: A microarray analysis. Br J Dermatol 2015;172:1555-61.
[Google Scholar]
|
| 40. |
Leirós GJ, Attorresi AI, Balañá ME. Hair follicle stem cell differentiation is inhibited through cross-talk between Wnt/β-catenin and androgen signalling in dermal papilla cells from patients with androgenetic alopecia. Br J Dermatol 2012;166:1035-42.
[Google Scholar]
|
| 41. |
Goldberg AD, Allis CD, Bernstein E. Epigenetics: A landscape takes shape. Cell 2007;128:635-8.
[Google Scholar]
|
| 42. |
Cobb JE, Wong NC, Yip LW, Martinick J, Bosnich R, Sinclair RD, et al. Evidence of increased DNA methylation of the androgen receptor gene in occipital hair follicles from men with androgenetic alopecia. Br J Dermatol 2011;165:210-3.
[Google Scholar]
|
Fulltext Views
34,705
PDF downloads
8,586





