Translate this page into:
Metagenomic next-generation sequencing for the aetiological diagnosis of Mycobacterium marinum infections: A pilot study
Corresponding author: Prof. Litao Zhang, Department of Dermatology, Tianjin Academy of Traditional Chinese Medicine Affiliated Hospital, Tianjin, China, zhanglitao@medmail.com.cn
-
Received: ,
Accepted: ,
How to cite this article: Gu A, Jiang J, Ma F, Zhang L. Metagenomic next-generation sequencing for the aetiological diagnosis of Mycobacterium marinum infections: A pilot study. Indian J Dermatol Venereol Leprol 2023;89:745-8
Dear Editor,
Skin infections caused by Mycobacterium marinum are very common in China. These infections present with clinically diverse manifestations, which makes it challenging for clinicians to diagnose and treat them. Metagenomic next-generation sequencing is currently being widely used to detect pathogens due to its fast identification, high accuracy and no culture requirement. 1 It has changed the traditional mode of pathogen diagnosis because of the quick identification of new and rare pathogens that can transmit cross-species and produce mixed infections. However, studies pertaining to the application of the metagenomic next-generation sequencing for the diagnosis of Mycobacterium marinum infections remain rare. Therefore, this study aimed to evaluate the application of metagenomic next-generation sequencing for diagnosing Mycobacterium marinum infection by comparing its efficacy with traditional pathogen detection methods (microbial culture, smear microscopy and histopathology).
All patients with a clinical diagnosis of Mycobacterium marinum infections were recruited retrospectively from Tianjin Academy of Traditional Chinese Medicine Affiliated Hospital from August 1st 2017 to May 31st 2021. Details of clinical pathological tests, smear microscopy, fungal and bacterial cultures and metagenomic next-generation sequencing data were retrieved and recorded. An incisional surgical biopsy was taken and part of the tissue was sent for routine hematoxylin-eosin, periodic acid-Schiff and acid-fast bacteria staining. The remaining sample was used for metagenomic next-generation sequencing pathogen detection. The deep tissue and pus of the skin lesions were also evaluated for smear microscopy by acid-fast and fluorescence staining, fungal culture and common bacterial and mycobacterial culture.
A total of 70 patients (22 males and 48 females) met the diagnostic criteria for Mycobacterium marinum infections, namely (1) patients with clear exposure to seafood or aquatic environments, especially with a positive history of puncture wounds, (2) patients with typical clinical manifestation, (3) patients with the pathological manifestations such as infectious granuloma without fungal hyphae or spores, with or without mycobacteria by acid-fast bacteria, (4) patients who had positive or negative mycobacterial cultures, but negative fungal cultures, (5) patients who received effective anti-mycobacterial antibiotic therapy.
Clinical data collected are enlisted in Table 1. The infection mainly involved skin and subcutaneous tissues leading to erythema, papules, nodules, plaques, ulcers, pustules and abscesses [Figures 1a to d].
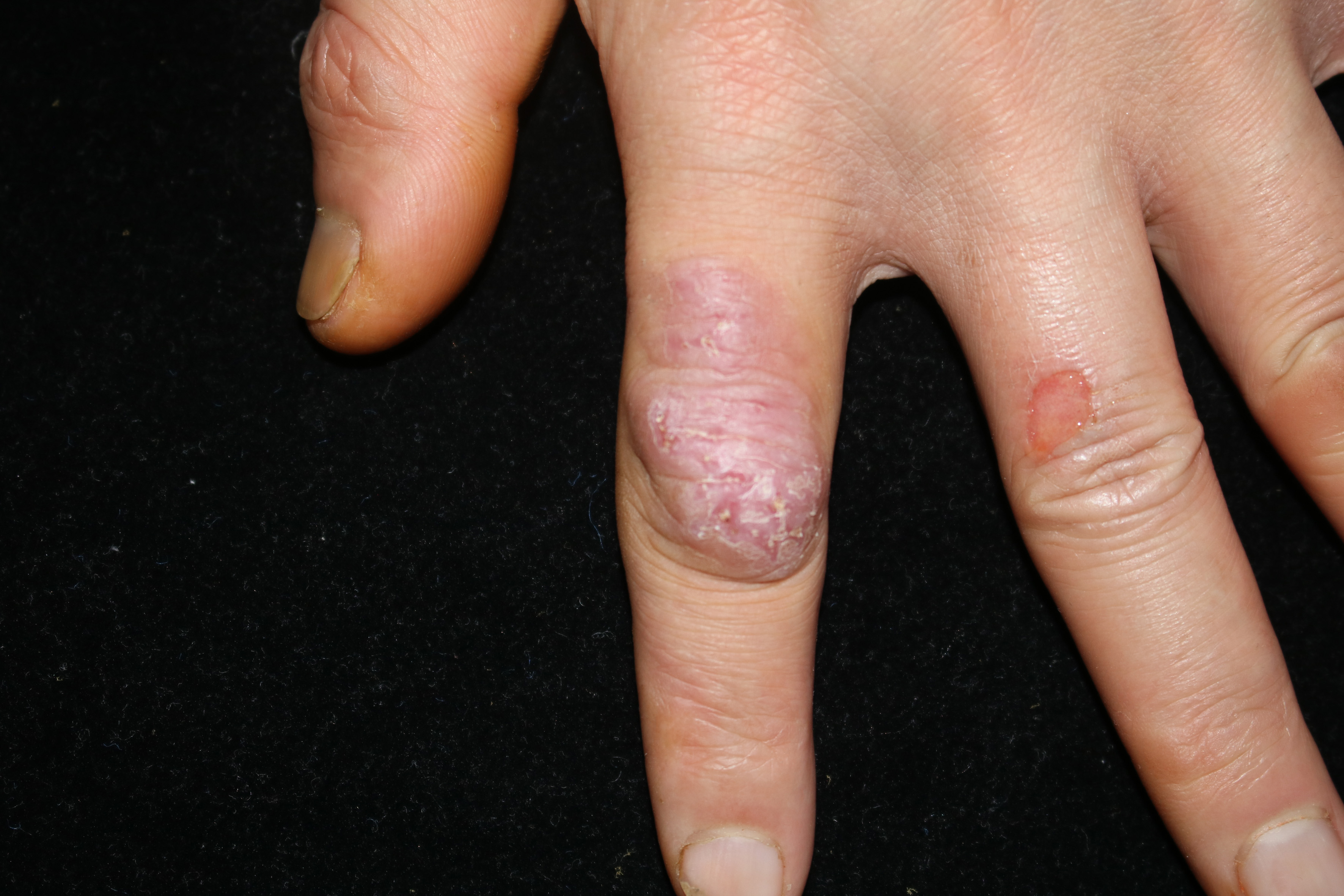
Single nodule
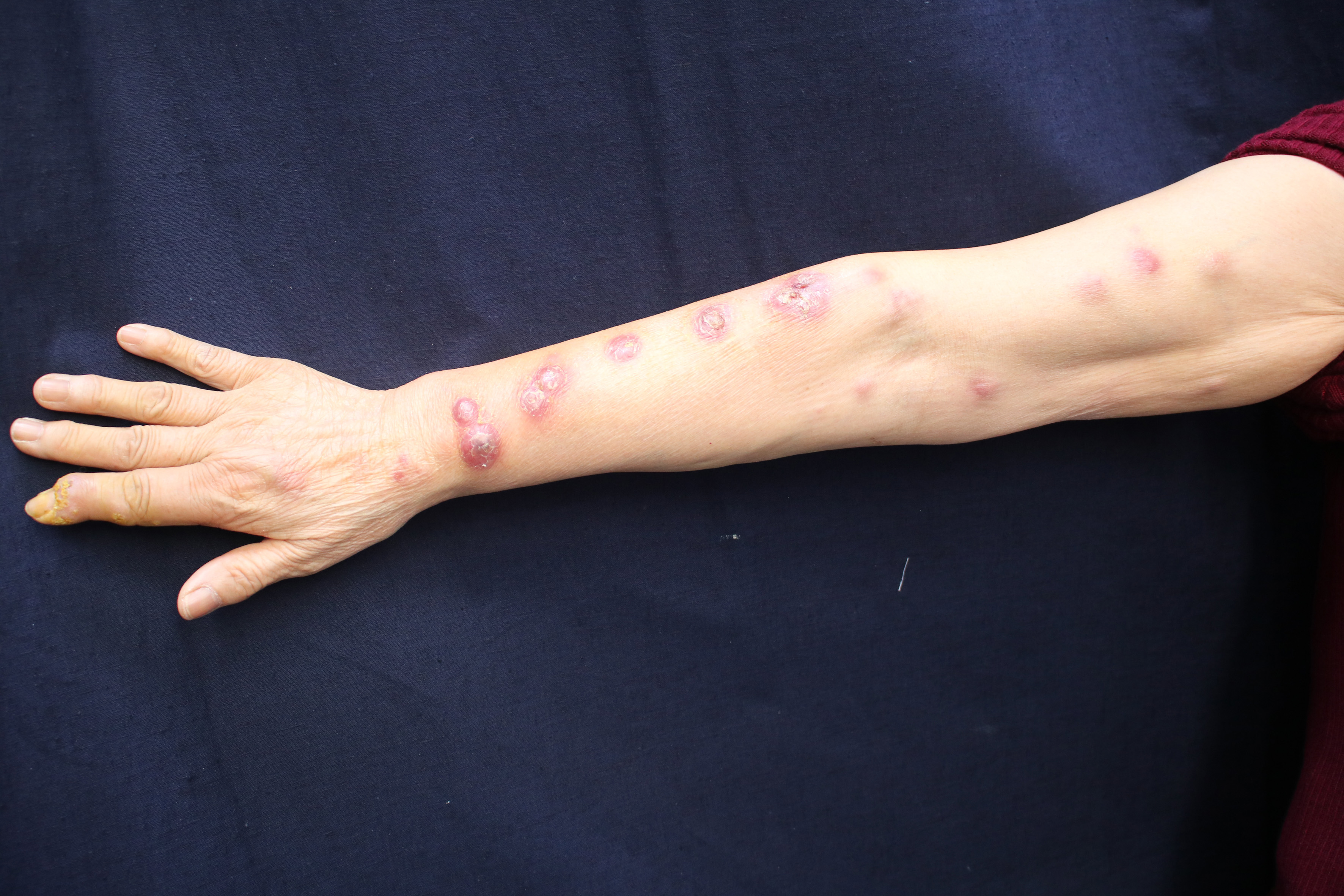
Unilateral beaded form
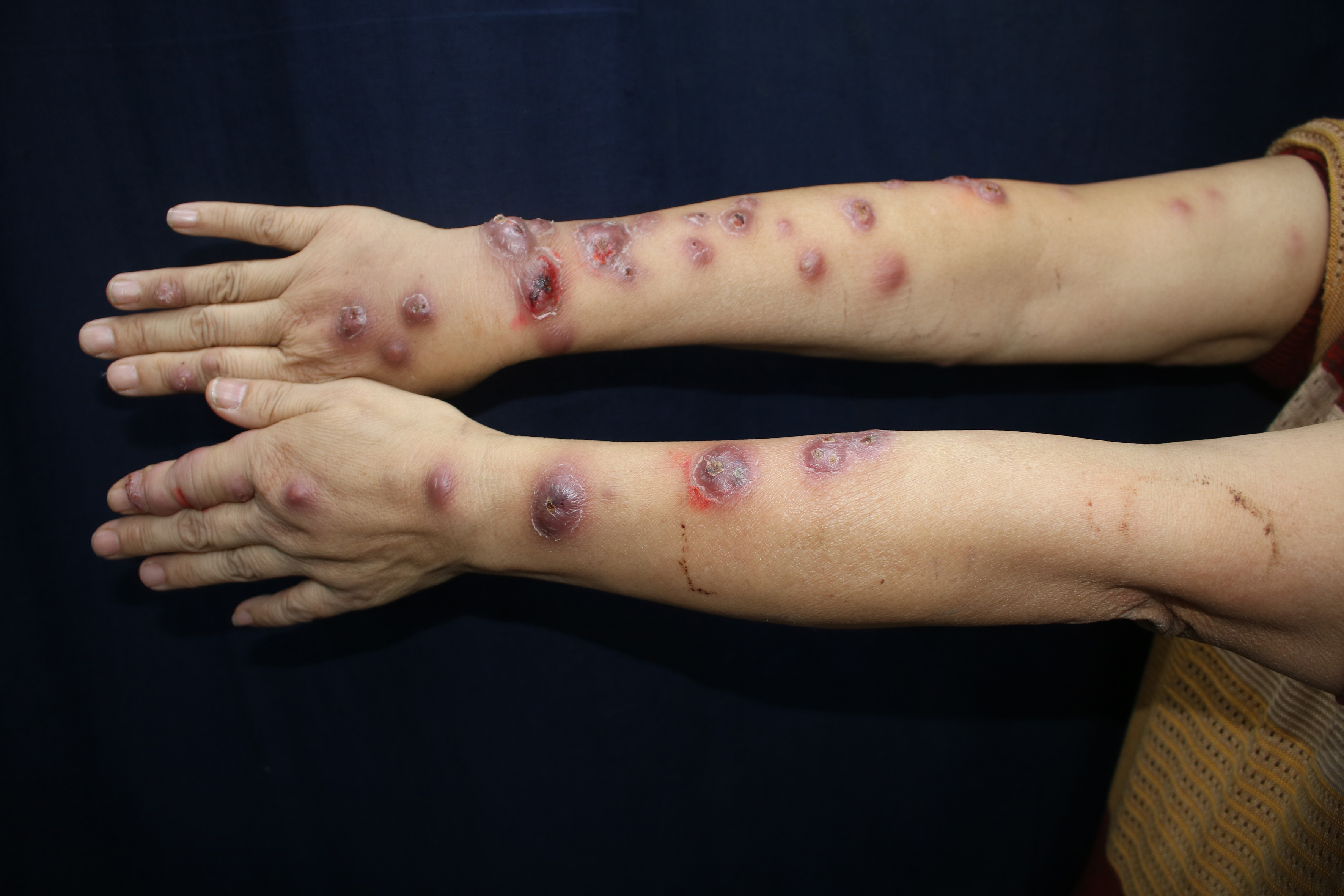
Bilateral beaded form
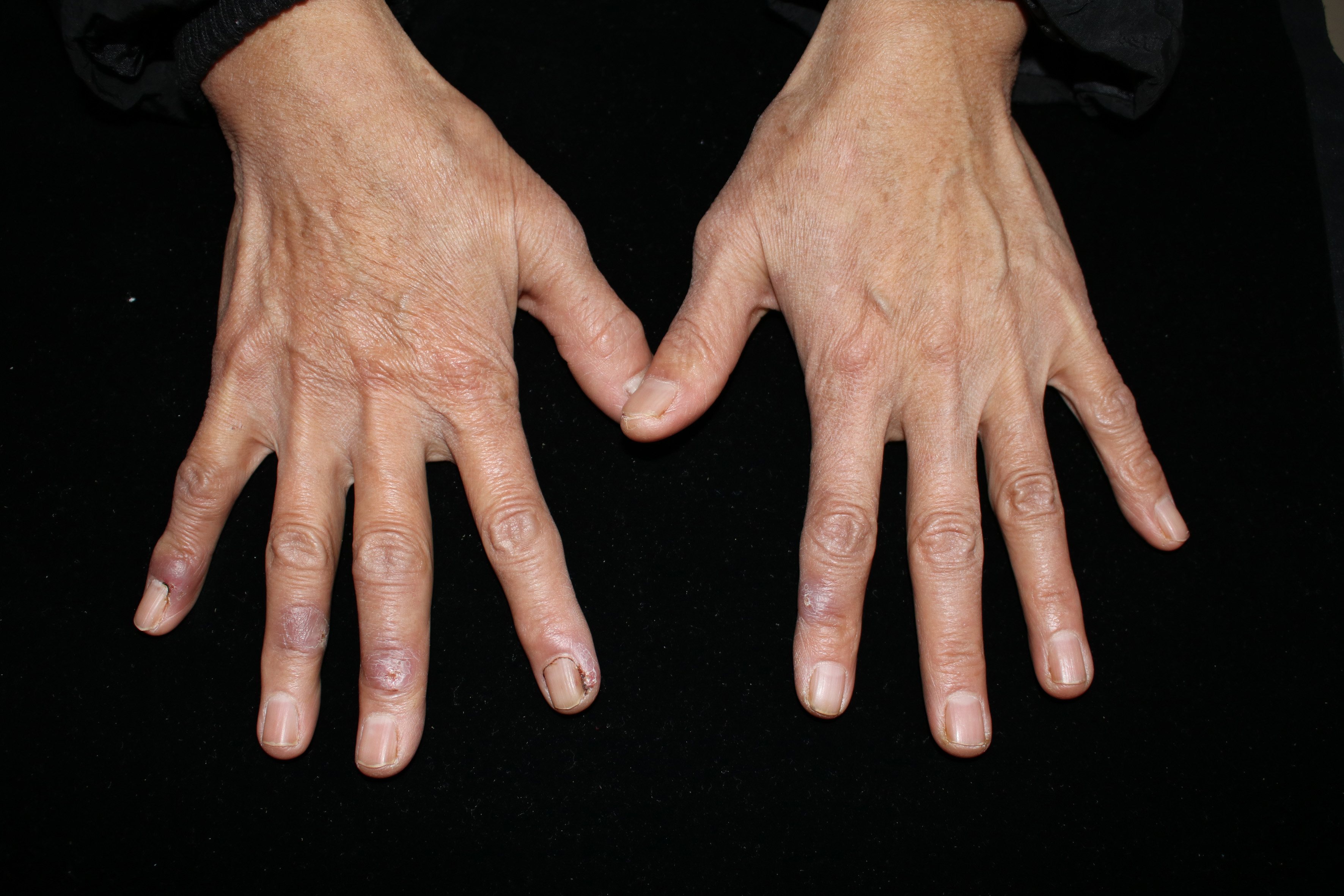
Bilateral scattered multiple type
Variables
Number (%)
Female
48 (68.6)
Age in years: mean; median
60.8; 62.0 (interquartile range: 38–79)
The incubation period in weeks: mean; median
4.6; 4.0 (interquartile range: 2–12)
Diagnosis time in a month: mean; median
3.4; 3.0 (interquartile range: 0.5–12 months)
ImmunocompromisedAntibiotics use
1 (1.4)68 (97.1)
History of aquatic exposure
Handled fish/seafood
63 (90.0)
Fish tank/aquarium
5 (7.1)
Fishing
2 (2.9)
Swimming/diving
0
Localization of infection
Finger/hand
24 (34.3)
Hand Involved arm
45 (64.3)
Involved only the forearm
1 (1.4)
Leg and truck
0
Distribution of the lesions
Unilateral single lesion
15 (21.4)
Unilateral beading form
36 (51.4)
Unilateral scattered multiple lesions
4 (5.7)
Bilateral beading form
9 (12.9)
Bilateral scattered multiple lesions
6 (8.6)
Treatment protocol and the outcome
Surgical excision for patients with a single lesion
4 (5.7)
Triple therapy with rifampicin, ethambutol and clarithromycin
completely disappeared after 12 weeks
14 (20.0)
completely disappeared after 24 weeks
16 (22.9)
completely disappeared after 18 months
11 (15.7)
Triple therapy combined with surgical resection
5 (7.1)
Transferred to a local tuberculosis hospital
20 (28.6)
The results of pus smear microscopy by acid-fast bacteria and fungal fluorescent staining of 70 patients were all negative. Laboratory culture and identification of deep tissue and pus revealed no bacterial growth after 3–4 days of culture and no fungal growth after 21 days of culture. However, 22 (31.4%) patients were positive for Mycobacterium marinum after 2–3 weeks of culture. Histologically, granulomas were detected in all samples, and mycobacteria were detected in 11 (15.7%) cases by the acid-fast bacteria method.
Mycobacterium marinum sequence was detected in 48 (68.6%) cases by metagenomic next-generation sequencing. After excluding the background bacteria, the number of matched nucleotide sequences ranged from 1 to 255, and the gene coverage was 0.001 ∼ 0.761%. Apart from Mycobacterium marinum, 20 cases in this study were associated with other pathogenic coinfections [Table 2]. The comparison of cost and positive rate between metagenomic next-generation sequencing and traditional detection methods is tabulated in Table 3.
Genus
Species
Number of cases
Number of sequences
Prokaryotes
Gram-positive bacilli
Corynebacterium
Corynebacterium falsenii
1
175
Staphylococcus
Staphylococcus epidermidis
1
120
Staphylococcus aureus
1
66
Mycobacterium
Mycolicibacterium smegmatis
1
1
Mycobacterium kansasii
1
1
Mycolicibacterium fortuitum
1
1
Mycobacterium Gordonae
1
1
Gram-negtive bacilli
Pseudomonas
Pseudomonas aeruginosa
1
313
Klebsiella
Klebsiella variicola
1
32
Raoultella
Raoultella ornithinolytica
1
255
Serratia
Serratia liquefaciens
1
100
Fugus
Candida
Candida parapsilosis
3
4246/2/25
Candida albicans
1
13
Saccharomyces
Saccharomyces cerevisiae
1
165
Virus
dsDNA
Lymphocryptovirus
Human gammaherpesvirus 4
3
13 /5/112
Roseolovirus
Human betaherpesvirus 7
1
3
Time taken
Cost ($)
Positive cases
Positivity (%)
Unintentional screening
Acid fast bacteria in smear microscopy
1 hour
1–2
0
0
No
Acid fast bacteria in histopathology
1 week
50–80
11
15.7
No
Microbial culture
2–3 weeks or longer
15–50
22
31.4
No
Metagenomic next-generation sequencing
2 days
200–400
48
68.6
Yes
Our study revealed that exposure to infected fish or seafood during handling or home cooking was the major risk factor for this infection in the Tianjin region, while cleaning fish tanks or aquarium maintenance accounted for a very small proportion, which is different from Holden’s report in 2018. 2 In the Tianjin region, Mycobacterium marinum preferentially affected elderly females with onset at a mean age of 60.8 years, these statistics differ from that in France and Denmark where middle-aged men were the most susceptible. 2 , 3 This may be because, in China, elderly females participate more in-home labor.
The bacterial load at the site of Mycobacterium marinum infection is very low making it difficult to find evidence by histology or acid-fast bacteria, whose positive rate in our study was 15.7%. Although the literature reported that 70–80% of Mycobacterium marinum infection cases were culture-positive, routine mycobacterial culture requires several weeks and reports from China had a lower positivity rate. 4 In this study, only 22 (31.4%) cases had positive culture results. Therefore, molecular biology is increasingly recognised as a faster and more sensitive diagnostic tool.
The metagenomic next-generation sequencing technology is an analysis and diagnosis system, which can objectively and directly detect a high-throughput nucleic acid sequence of pathogenic micro-organisms (including viruses, bacteria, fungi, parasites, etc.) in clinical samples, and then compare it with the database for analysis. 5 The sequence does not need specific amplification and is not affected by antibiotics. In this study, metagenomic next-generation sequencing detected the majority of Mycobacterium marinum cases by nucleic acid sequence even after multiple antibiotics were used; its sensitivity was confirmed as in nine cases, it only detected one nucleic acid fragment. Its positive rate reached 68.6%, which is significantly higher than histological acid-fast staining, smear microscopy and tissue culture. Additionally, the metagenomic next-generation sequencing detection of 20 cases revealed the presence of other nonpathogenic coinfection, including Gram-positive and negative bacteria, fungi and dsDNA viruses. Compared with traditional detection methods, which can only conduct intentional screening and can’t achieve wide coverage of pathogen inspection, metagenomic next-generation sequencing can simultaneously exclude and identify other pathogenic infections in a short period. 1
Our study has inherent limitations: the true incidence of Mycobacterium marinum infections might be greater than estimated in this study, as we included only patients with histopathological and etiological tests, and patients treated based on a clinical diagnosis were left out. Therefore, more studies are required to reach definitive conclusions.
We believe that metagenomic next-generation sequencing is an effective tool for the early detection and control of Mycobacterium marinum infection and can provide valuable information allowing individualised and optimised antibiotic treatment of patients. However, this test also has its own shortcomings, such as a lack of bacterial subculture and drug susceptibility and higher costs. So traditional detection methods, especially microbial culture and acid-fast bacteria in histopathology, still remain popular. The combined detection can be considered if the patient’s financial situation permits.
Declaration of patient consent
The authors certify that they have obtained all appropriate patient consent.
Financial support and sponsorship
Nil.
Conflict of interest
There are no conflicts of interest.
References
- Clinical metagenomic next-generation sequencing for pathogen detection. Annu Rev Pathol. 2019;14:319-38.
- [CrossRef] [PubMed] [Google Scholar]
- Mycobacterium marinum infections in Denmark from 2004 to 2017: A retrospective study of incidence, patient characteristics, treatment regimens and outcome. Sci Rep. 2018;8:6738.
- [Google Scholar]
- Sixty-three cases of Mycobacterium marinum infection: Clinical features, treatment, and antibiotic susceptibility of causative isolates. Arch Intern Med. 2002;162:1746-52.
- [PubMed] [Google Scholar]
- Mycobacterium marinum. Microbiol Spectr. 2017;5 doi: 10.1128/microbiolspec.TNMI7-0038-2016
- [CrossRef] [PubMed] [Google Scholar]
- Metagenomic next-generation sequencing versus traditional pathogen detection in the diagnosis of peripheral pulmonary infectious lesions. Infect Drug Resist. 2020;13:567-76.
- [Google Scholar]





