Translate this page into:
Report of rpoB mutation in clinically suspected cases of drug resistant leprosy: A study from Eastern India
2 Department of Dermatology, Calcutta School of Tropical Medicine, Kolkata, West Bengal, India
3 Stanley Browne Research Laboratory, The Leprosy Mission, New Delhi, India
4 Department of Biochemistry, B. S Medical College, Bankura, West Bengal, India
5 National JALMA Institute for Leprosy and Other Mycobacterial Diseases, Agra, Uttar Pradesh, India
6 Department of Biochemistry, Pt. N. M. Medical College, Raipur, Chhattisgarh, India
Correspondence Address:
Basudev Bhattacharya
Department of Biochemistry, Institute of Post Graduate Medical Education and Research, Kolkata, West Bengal
India
| How to cite this article: Hasanoor Reja AH, Biswas N, Biswas S, Lavania M, Chaitanya VS, Banerjee S, Maha Patra PS, Gupta UD, Patra PK, Sengupta U, Bhattacharya B. Report of rpoB mutation in clinically suspected cases of drug resistant leprosy: A study from Eastern India. Indian J Dermatol Venereol Leprol 2015;81:155-161 |
Abstract
Background: The current strategy for leprosy control depends mainly on early case detection and providing the recommended multidrug therapy (MDT) dosage. Understanding the molecular mechanisms of drug resistance to each of these drugs is essential in providing effective treatment and preventing the spread of resistant strains in the community. The progress of molecular biology research provides a very efficient opportunity for the diagnosis of drug resistance by in vitro method. Aim: We aimed to investigate the point mutations within the rpoB gene region of the Mycobacterium leprae genome, which are responsible for resistance to rifampicin, in order to determine the emergence of drug resistance in leprosy in the Kolkata region of West Bengal. Methods: A total of 50 patients with a relapse of leprosy were enrolled in the study. Skin smears were obtained for estimation of bacillary index and biopsies were obtained in 70% alcohol for extraction of DNA. The extracted DNA was amplified by M. leprae-polymerase chain reaction (PCR) targeting rpoB gene region. Every single nucleotide base in the sequence is aligned to reference sequence and identity gaps were determined by NCBI - BLAST. Later in-silico analysis was done to identify the changes in the translated protein sequences. Results: A mutation at the base pair position 2275405 where G is replaced by C in the M. leprae genome, which corresponds to the coding region of rpoB gene (279 bp - 2275228 to2275506), was observed in two patients. This missense mutation in CAC codon brings about a glutamic acid to histidine change in the amino acid sequence of RNA polymerase beta subunit at the position 442 (Glu442His), a region specific for rifampicin interaction, which might be responsible for unresponsiveness to rifampicin by manifesting a stable bacteriological index in these 2 patients even after completion of 24 months of multibacillary multi-drug therapy (MB-MDT). Limitations: The major limitations of multiple-primer PCR amplification refractory mutation system (MARS) assay is that it capable of detecting mutation at codon 425 and cannot distinguish any silent amino acid changes. Conclusion: The study indicates the existence of rifampicin drug resistance in Eastern India.INTRODUCTION
Drug resistance in leprosy may be a serious impediment during the present scenario of dramatic decline in the prevalence rate to <1/10,000, which has been attained throughout the world due to implementation of intensive and concerted chemotherapeutic approach of multidrug therapy (MDT) by the World Health Organization (WHO). Until 1982, the standard treatment for lepromatous leprosy was lifelong monotherapy with dapsone or till attainment of bacterial negativity, which resulted in the development of dapsone resistant Mycobacterium leprae (M. leprae) leading to its therapeutic failure. Multidrug therapy with dapsone, and rifampicin, with or without clofazamine, recommended by the WHO in 1982 was implemented in 1985 throughout the world and includes rifampicin, the only bactericidal drug against M. leprae. However, rifampicin-resistant leprosy has been documented in patients treated by rifampicin monotherapy. [1],[2],[3] The emergence of multidrug resistant M. leprae strains, which has been reported from South East Asia, [4] indicates a serious threat for control of leprosy. Unlike M. tuberculosis, M. leprae is a slow growing bacterium and has not yet been successfully cultured in vitro. Thus, identification of drug-resistant M. leprae is challenging. An in vivo model, the mouse foot pad (MFP) method of Shepard has been utilized for drug susceptibility testing. [5] Briefly, biopsies containg M. leprae are obtained from skin lesions in patients and inoculated into the mouse foot pad. The mice are treated with anti-M. leprae drugs to determine whether the drug prevents bacterial multiplication in the foot pad. [6] This method requires a large biopsy with a yield of at least 5 × 10 4 organisms which should be inoculated within 72 hours, is time consuming needing a minimum of 6 months to obtain results and requires expensive facilities and expertise, that are not easily available in most laboratories. Recently, point mutations in M. leprae genome responsible for dapsone and rifampicin resistance have been identified, which has facilitated the development of new tools for monitoring the drug resistance. [7],[8],[9] Rifampin/Rifampicin, (3-{[4-met hyl-1-pi perazinyl0-imino]-methyl} rifamycin) is the key bactericidal component of the MDT regimen. A single dose of 1200 mg can reduce the number of viable bacilli in skin to undetectable levels within a few days. [10] The target for rifampicin in M. leprae is the β subunit of RNA polymerase encoded by rpoB gene. [11] Mycobacterial resistance to rifampicin correlates with changes in the structure of the subunit of the DNA-dependent RNA polymerase, primarily due to missense mutations within codons of a highly conserved region of the rpoB gene referred to as the rifampicin resistance-determining region. [12] Substitution within codon Ser-425-Leu has shown to be the most frequent mutation associated with the development of the rifampicin-resistant phenotype in M. leprae. [13],[14],[15] In the recent past, utilizing various species specific primers, our group has developed a multiplex PCR for diagnosis of leprosy. [16] In the present study, we analyzed the point mutations within the rpoB gene region of M. leprae, which are responsible for resistance to rifampicin, in order to determine the emergence of drug resistance in leprosy in the Kolkata region of West Bengal.
METHODS
The study was conducted in the department of biochemistry at the Institute of Post Graduate Medical Education and Research, Kolkata from July 2007 to July 2009. The study primarily included patients attending the out patient department (OPD) of the department of dermatology. We also collected a few samples from dermatology OPDs of other medical colleges of Kolkata and also from different leprosy clinics of West Bengal. Each patient was informed in detail about the study in the local language and duly signed written informed consent was obtained from every patient before their enrolment in the study. The study commenced after obtaining clearance from the ethical committee of the Institution.
Selection criteria for patients
- Patients classified as either paucibacillary (PB) or multibacillary (MB) leprosy, with incomplete or irregular therapy who relapsed with increase in bacteriological index (BI)
- Patients with appearance of new skin patches or nodules or new nerve involvement after completion of MDT and with increase in BI of 1 or more units at any single site were considered as relapse patients.
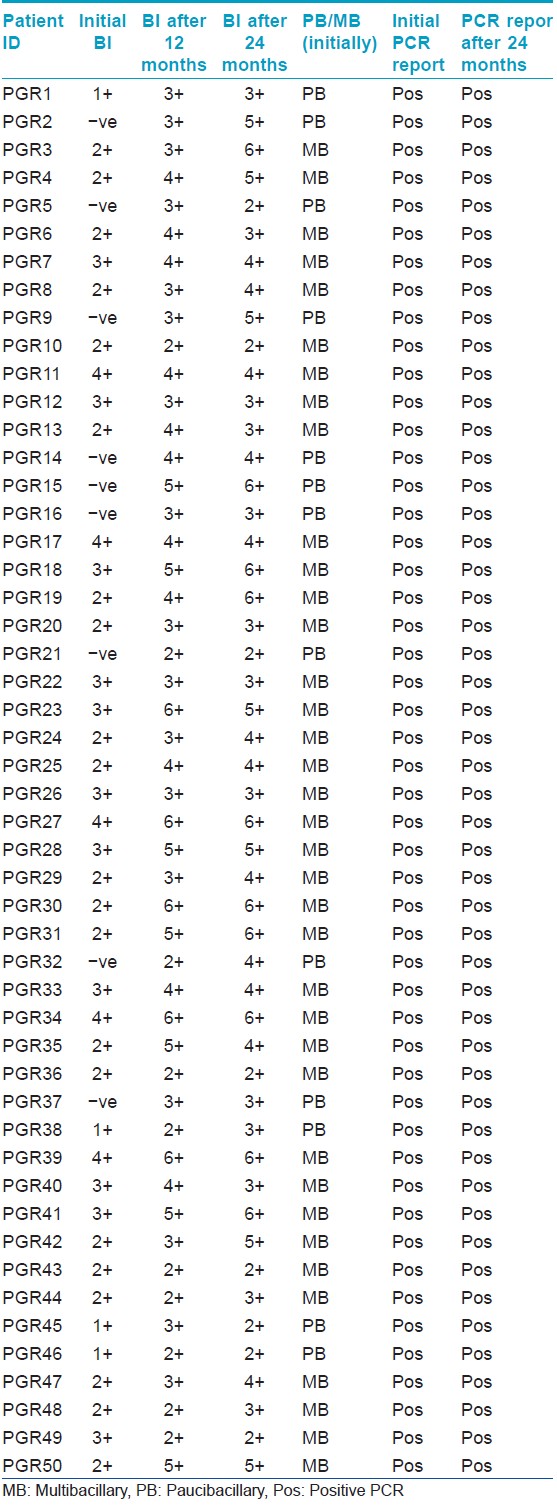
A total of 50 cases of the above-mentioned categories attending the OPD were enrolled as shown in [Table - 1]. Since facility for the mouse foot pad assay is not available at our institute, the known wild-type Thai 53 strain [7] (obtained from Louisiana State University, USA) was included as control.
Sample collection
- Slit skin smears samples were obtained from the edge of the most active lesions and were stained by Ziehl-Neelsen staining method [16] to determine the bacillary index (BI)
- Punch (4 mm) biopsies obtained from the active lesions were collected in 70% ethanol and processed for DNA extraction according to our earlier described method. [16]
Extraction of genomic DNA from biopsies
The skin biopsy samples were homogenized in 100 μl of lysis buffer (containing 100Mm Tris-HCl, 0.05% Tween-20 and 60 mg/ml of Proteinase-K, pH 8.5) using a hand-operated homogenizer (Z35997-1; Sigma) with a plastic pestle, and then incubated for 18 h at 60°C after mineral oil was overlaid on the upper surface of the sample to avoid evaporation. This was followed by the inactivation of proteinase K at 95°C for 15 min. An equal volume of phenol/chloroform/isoamyl alcohol (25:24:1) was added to the samples, mixed by vortexing for 30 s, and then centrifuged at 10,000 ×g for 15 min at 4°C. The aqueous phase was transferred to a clean tube, and the procedure was repeated with phenol-chloroform-isoamylalcohol (25:24:1) until a clear interface was observed. A final extraction with chloroform-isoamylalcohol (24:1) was performed and after centrifugation, aqueous layer was collected. To the aqueous layer, isopropanol (0.7% of the total volume of the sample) was added and incubated overnight at -20°C. The samples were then centrifuged at 12,000 rpm for 20 min, the supernatant was removed, and the DNA pellet was washed with ice-cold 70% ethanol. The DNA pellet was then allowed to air dry and was resuspended in 100 μl of sterile distilled water, which was subsequently used as a template DNA for polymerase chain reaction (PCR).
Detection of M.leprae DNA by multiplex PCR
The extracted DNA was amplified by multiplex PCR developed in our laboratory for diagnosis of M. leprae. The primers R1 and R2 and TTC-A and TTC-B were used and the amplicons were electrophoresed on a 2% agarose gel to determine the presence of M. leprae DNA as described by us earlier. [16] Later, the DNA samples were processed for the detection of rpoB gene mutation responsible for rifampicin resistance.
Detection of Mutation in rpoB Gene
The rpoB gene was analyzed by direct sequencing of PCR amplified products generated using conditions mentioned by WHO guidelines for Global Surveillance of Drug Resistance in Leprosy. [17] The primers used were OMSrpoBF GTCGAGGCGATCACGCCGCA and OMSrpoBR CGACAATGAACCGATCAGAC which generated a PCR product of 279 bp. The PCR reaction mix contains 12.5 μl PCR master mix (2×) containing Hot Start Taq polymerase, from Qiagen (Qiagen, USA), both primers at final volume of 0.5 μl (0.25 μM), approximately 2 μl (2 μg) of template DNA and nuclease free water at a final volume of 4.5 μl. PCR was carried out in a Thermocycler (Hybaid Inc. USA) and the cycling conditions included 1 cycle of initial denaturation at 95°C for 15 min followed by 40 cycles of denaturation at 94°C for 1 min, annealing at 61°C for 1 min and extension at 72°C for 1 min and a single cycle of final extension at 72°C for 10 min. After amplification, the amplicons were loaded in 2% agarose gel. The mutation was further confirmed by DNA sequencing. PCR products were outsourced for commercial sequencing (Xplorigen Technologies Pvt Limited, Delhi, India). Sequence data was analysed by using Molecular Evolutionary Genetics Analysis (MEGA) (http://www.megasoftware.net/) version 5.
RESULTS
RpoB gene was amplified in 50 samples chosen for the study. Fifteen microliter of each amplicon was electrophoresed on a 2% agarose gel to determine the presence of band of 279 bp, which was visually identified using molecular band size markers (Alpha Imager Inc. USA) and by using a Step up TM 100 bp DNA ladder (Bangalore Genei). Later the bands were cut on a manual UV Illuminator and eluted using Gel Elution Kit (Sigma Aldrich Inc.) as shown in [Figure - 1]. Fifteen micro liter of the eluted DNA along with 10 μM forward primer was sent to Xplorigen Inc. India for DNA sequencing. The sequence data in the form of ABI files as shown in [Figure - 2] were analysed using MEGA 5 Software. DNA sequence was compared to a known database of M. leprae genome to determine similarity and identity percentage using Basic Local Alignment Search Tool (BLAST) and sites where there was no identity were noted. Later the sequence was translated online using http://insilico.ehu.es/translate/into a first reading frame amino acid sequence, which was again compared with known amino acid sequence from the NCBI database using BLASTp (Protein BLAST).
 |
| Figure 1: 2% Agarose Gel Electrophoresis Image showing 279 bp band corresponding to PCR-based amplification of rpoB gene |
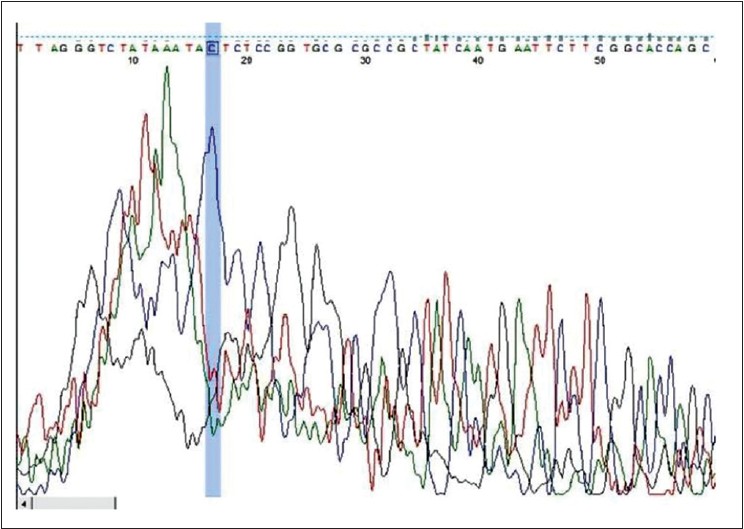 |
| Figure 2: ABI File (Chromatogram) with point mutation |
Nucleotide BLAST Report indicating a base pair change at 2275405 loci.

Our results revealed a mutation at the base pair position 2275405 where G is replaced by C in the M. leprae genome, which corresponds to the coding region of rpoB gene (279 bp -2275228 to 2275506). This is a nonsynonymous missense mutation in CAC codon, which brings about a glutamic acid to histidine change in the amino acid sequence of RNA polymerase beta subunit of M. leprae at position 442 (Glu442His), a region specific for rifampicin interaction. This mutation was observed in 2 out of the 50 patients. Clinically these patients demonstrated unresponsiveness to rifampicin by manifesting no reduction in bacterial load as measured by bacillary index (BI) even after completion of 24 months of multibacillary multidrug therapy (MB MDT).
DISCUSSION
The current strategy for leprosy control depends mainly on early case detection and providing the recommended MDT dosage. Understanding the molecular mechanisms of drug resistance to each of these drugs is essential in providing effective treatment and preventing the spread of resistant strains in the community. M. leprae drug resistance was reported in 1964 for dapsone, [18] in 1993 for rifampin, [12] and in 2000 for ofloxacin. [14] Discontinuation of treatment and monotherapy play a major role in development of drug resistant strains of M. leprae. In leprosy, drug susceptibility is measured by cure of clinical symptoms and reduction in bacterial load. However, patients who relapse or whose bacillary index (BI) does not fall even after administration of multidrug therapy (MDT) should be screened for drug resistance and the most appropriate antibiotic regimen needs to be initiated. In addition, such screening is the only way to measure the epidemiological trend of primary or secondary resistance to anti-leprosy drugs. [19] Drug susceptibility testing by the mouse foot pad method is time consuming and expensive. The progress of molecular biology research provides a very efficient opportunity for the diagnosis of drug resistance by in vitro method. [20] In this study, a total of 50 cases were monitored for the detection of mutation in rpoB gene, which is responsible for drug resistance against rifampicin. We observed 2 out of 50 study subjects to be resistant to rifampicin. These patients had relapsed with new active skin lesions even after completion of multidrug therapy (MDT). Since mouse foot pad assay was not available in our centers, we compared our results using a molecular method by simultaneous comparison with the known wild strain of leprosy (Thai53) and further confirmed the results by sequencing.
Rifampicin is a key drug in the chemotherapeutic regimen used to combat both leprosy and tuberculosis. Owing to its efficient bactericidal properties against M. leprae, this drug remains the backbone of the multidrug therapy (MDT) regimen. It is conceivable, however, that rifampicin resistance could compromise the control program and owing to the very slow growth rate of M. leprae, it may take a long time to appear in the population at large. Therefore, at this stage there is a real need for rapid and efficient tools for determining whether drug-resistant M. leprae is on the horizon. An earlier search [21] in south India reported M. leprae resistance to dapsone but was unable to detect any molecular evidence of rifampicin resistance. The PCR-based assay convincingly demonstrated that detection of rifampicin resistance, is a feasible and practical alternative assay over the mouse foot pad assay and has application in developing countries where the leprosy burden is relatively high. Although a major limitation to molecular genetic determination of drug resistance by any technique is that molecular genetic tests detect only known mutations, it provides a rapid screening tool for the majority of resistant isolates, which, in turn, allows a reduction in the amount of phenotypic drug susceptibility testing.
A major limitation of the multiple-primer PCR amplification refractory mutation system (MARS) assay as it is currently designed is that it is only capable of detecting mutations at codon 425 in the rpoB gene of M. leprae, and does not detect other mutations known to be associated with rifampicin resistance, or indeed other potential mutations that could cause resistance. Furthermore, as this test detects only nucleotide mutations, it cannot distinguish silent amino acid changes from those that result in amino acid substitution, although no silent mutation has been reported to date at the codon 425 position in the rpoB gene of M. leprae.[22] However, we justified our result by amplifying the whole Rifampicin Resistance Determining Region (RRDR) followed by single-strand conformation polymorphism (SSCP) to confirm that no unknown mutation exists. We were unable to find previous reports of mutation/s that confer rifampicin resistance as noted in two of the patients in our study.
Further work needs to be carried out to establish the presence of this phenomenon in this part of India and elsewhere. Though we have already achieved and declared a prevalence rate of <1/10,000 and eliminated leprosy as a public health problem, this report is significant in terms of remaining challenges and the way forward toward final and complete elimination. The moderate efficiency of the in vivo mouse foot pad-based method for susceptibility testing compared with its cost and the organizational need to maintain the mouse foot pad test facilities puts in doubt the future viability of such a technique for determination of M. leprae resistant strains. Nevertheless, it may be premature to abandon the test because of the relatively small number of cases of leprosy analyzed worldwide using the newer molecular techniques. However, understanding the magnitude of drug resistance and keeping current MDT effective while monitoring the drug resistance cases carefully is essential to prevent the spread of drug resistance.
CONCLUSION
Drug susceptibility testing is necessary for leprosy patients with relapse or whose symptoms do not improve in spite of adequate therapy with multidrug therapy (MDT). Emergence of drug resistance would create tremendous difficulty for the treatment of individual patients, and its spread would pose a serious threat in achieving the final goal of elimination. The study indicates the existence of rifampicin drug resistance in eastern India. Since this is to our knowledge the first report of its kind from this region, it suggests that drug resistance surveillance is essential to keep a vigil on the drug resistance scenario at many vulnerable settings in spite of the declining leprosy trend and thereby attaining the success of the final goal of leprosy elimination as a public health problem.
| 1. |
Jacobson RR, Hastings RC. Rifampin-resistant leprosy. Lancet 1976;2:1304-5.
[Google Scholar]
|
| 2. |
Grosset JH, Guelpa-Lauras CC, Bobin P, Brucker G, Cartel JL, Constant-Desportes M, et al. Study of 39 documented relapses of multibacillary leprosy after treatment with rifampin. Int J Lepr Other Mycobact Dis 1989;57:607-14.
[Google Scholar]
|
| 3. |
Cambau E, Bonnafous P, Perani E, Sougakoff W, Ji B, Jarlier V. Molecular detection of rifampin and ofloxacin resistance for patients who experience relapse of multibacillary leprosy. Clin Infect Dis 2002;34:39-45.
[Google Scholar]
|
| 4. |
Kai M, Nguyen Phuc NH, Nguyen HA, Pham TH, Nguyen KH, Miyamoto Y, et al. Analysis of drug-resistant strains of Mycobacterium leprae in an endemic area of Vietnam. Clin Infect Dis 2011;52:e127-32.
[Google Scholar]
|
| 5. |
Shepard CC. The experimental disease that follows the injection of human leprosy bacilli into foot-pads of mice. J Exp Med 1960;112:445-54.
[Google Scholar]
|
| 6. |
Shepard CC. A survey of the drugs with activity against M. leprae in mice. Int J Lepr Other Mycobact Dis 1971;39:340-8.
[Google Scholar]
|
| 7. |
Williams DL, Spring L, Harris E, Roche P, Gillis TP. Dihydropteroate synthase of Mycobacterium leprae and dapsone resistance. Antimicrob Agents Chemother 2000;44:1530-7.
[Google Scholar]
|
| 8. |
Kai M. [Diaminodiphenylsulfone resistance of Mycobacterium leprae due to mutations in the dihydropteroate synthase gene]. Nihon Hansenbyo Gakkai zasshi 2004;73:221-6.
[Google Scholar]
|
| 9. |
Cambau E, Carthagena L, Chauffour A, Ji B, Jarlier V. Dihydropteroate synthase mutations in the folP1 gene predict dapsone resistance in relapsed cases of leprosy. Clin Infect Dis 2006;42:238-41.
[Google Scholar]
|
| 10. |
Levy L, Shepard CC, Fasal P. The bactericidal effect of rifampicin on M. leprae in man: (a) Single doses of 600, 900 and 1200 mg; and (b) daily doses of 300 mg. Int J Lepr Other Mycobact Dis 1976;44:183-7.
[Google Scholar]
|
| 11. |
Zhang L, Namisato M, Matsuoka M. A mutation at codon 516 in the rpoB gene of Mycobacterium leprae confers resistance to rifampin. Int J Lepr Other Mycobact Dis 2004;72:468-72.
[Google Scholar]
|
| 12. |
Honore N, Cole ST. Molecular basis of rifampin resistance in Mycobacterium leprae. Antimicrob Agents Chemother 1993;37:414-8.
[Google Scholar]
|
| 13. |
Cambau E, Perani E, Guillemin I, Jamet P, Ji B. Multidrug-resistance to dapsone, rifampicin, and ofloxacin in Mycobacterium leprae. Lancet 1997;349:103-4.
[Google Scholar]
|
| 14. |
Matsuoka M, Kashiwabara Y, Namisato M. A Mycobacterium leprae isolate resistant to dapsone, rifampin, ofloxacin and sparfloxacin. Int J Lepr Other Mycobact Dis 2000;68:452-5.
[Google Scholar]
|
| 15. |
Williams DL, Gillis TP. Molecular detection of drug resistance in Mycobacterium leprae. Lepr Rev 2004;75:118-30.
[Google Scholar]
|
| 16. |
Banerjee S, Ray D, Bandyopadhyay D, Gupta S, Gupta S, Ghosal C, et al. Development and application of a new efficient and sensitive multiplex polymerase chain reaction (PCR) in diagnosis of leprosy. J Indian Med Assoc 2008;106:436-40.
[Google Scholar]
|
| 17. |
Asia WROf SE. Guidelines for global surveillance of drug resistance in Leprosy. WHO Leprosy 2009:32.
[Google Scholar]
|
| 18. |
Pettit JH, Rees RJ. Sulphone resistance in leprosy. An experimental and clinical study. Lancet 1964;2:673-4.
[Google Scholar]
|
| 19. |
Gupta UD, Katoch K, Singh HB, Natrajan M, Katoch VM. Persister studies in leprosy patients after multi-drug treatment. Int J Lepr Other Mycobact Dis 2005;73:100-4.
[Google Scholar]
|
| 20. |
Sekar B, Arunagiri K, Kumar BN, Narayanan S, Menaka K, Oommen PK. Detection of mutations in folp1, rpoB and gyrA genes of M. leprae by PCR-direct sequencing-A rapid tool for screening drug resistance in leprosy. Lepr Rev 2011;82:36-45.
[Google Scholar]
|
| 21. |
Shepard CC. A kinetic method for the study of activity of drugs against Mycobacterium leprae in mice. Int J Lepr 1967;35: 429-30.
[Google Scholar]
|
| 22. |
Sapkota BR, Ranjit C, Neupane KD, Macdonald M. Development and evaluation of a novel multiple-primer PCR amplification refractory mutation system for the rapid detection of mutations conferring rifampicin resistance in codon 425 of the rpoB gene of Mycobacterium leprae. J Med Microbiol 2008;57:179-84.
[Google Scholar]
|
Fulltext Views
3,873
PDF downloads
2,013





