Translate this page into:
Critical role of epigenetic modification in the pathogenesis of atopic dermatitis
Corresponding author: Prof. Jianyun Lu, Department of Dermatology, the Third Xiangya Hospital, Central South University, Changsha, Hunan, China. xiaoyun3@csu.edu.cn
-
Received: ,
Accepted: ,
How to cite this article: Chen C, Zeng J, Lu J. Critical role of epigenetic modification in the pathogenesis of atopic dermatitis. Indian J Dermatol Venereol Leprol 2023;89:700-9
Abstract
Atopic dermatitis is a chronic inflammatory skin disease characterised by recurrent eczema-like lesions and severe pruritus, along with drying and decrustation of skin. Current research relates the pathogenesis of atopic dermatitis mainly to genetic susceptibility, abnormal skin barrier function, immune disorders, Staphylococcus aureus colonisation, microbiological dysfunction and vitamin D insufficiency. Epigenetic modifications are distinct genetic phenotypes resulting from environment-driven changes in chromosome functions in the absence of nuclear DNA sequence variation. Classic epigenetic events include DNA methylation, histone protein modifications and non-coding RNA regulation. Increasing evidence has indicated that epigenetic events are involved in the pathogenesis of atopic dermatitis by their effects on multiple signalling pathways which in turn influence the above factors. This review primarily analyses the function of epigenetic regulation in the pathogenesis of atopic dermatitis. In addition, it tries to make recommendations for personalised epigenetic treatment strategies for atopic dermatitis in the future.
Keywords
Atopic dermatitis
DNA methylation
histone modification
non-coding RNA
pathogenesis
Introduction
Atopic dermatitis (AD) is a common chronic inflammatory skin disease characterised by dry skin and eczema-like lesions, accompanied by persistent itching.
The disease has a prolonged waxing and waning course which can potentially have a severe impact on patients’ quality of life.1 Admittedly, the pathophysiology of AD is very varied and complicated involving systemic immune dysfunction driven by T helper cells 2 (Th2 cells) and keratinocyte (KC) dysfunction. The mechanisms of AD are unclear and it is believed to be a multifactorial genetic disease induced by epithelial barrier dysfunction, abnormal innate and adaptive immune responses, S. aureus colonisation, intestinal and cutaneous microbial dysbiosis, etc.2 These mechanisms are mutually interlinked and create a vicious cycle eventually. More and more evidence indicates that modern lifestyles3,4 (i.e., excessive hygiene, western diet) and other environmental factors5,6 such as pollution and passive smoking also regulate susceptibility to AD. However, we still have a limited understanding of the potential mechanisms of the increased morbidity of AD mediated by environmental exposures. In this scenario, epigenetics provides a novel explanation for gene-environment interactions.
Epigenetics refers to functionally relevant changes in a chromosome without alterations in the DNA sequence, which has an important contribution to phenotypic plasticity. It mainly encompasses CpG island DNA methylation, histone modification and non-coding RNA-mediated regulation. Epigenetic events often have their own unique autogenetic trigger which is susceptible to various environmental factors.7 Epigenetic modifications influence various biological processes, especially cell proliferation and differentiation. Therefore, increasing evidence suggests that abnormal epigenetic modifications contribute to the pathophysiology of AD.8,9 For example, both recombinant keratin 6A (KRT6A) and KRT6B were overexpressed in AD lesions, which was related to the reduction of single CpG methylation in KRT6A. Further research demonstrated that miR-143 inhibited IL-13-induced downregulation of epidermal barrier-related proteins (filaggrin, loricrin, etc.) and inflammation by targeting IL-13 receptors.10
This review focuses on the contribution of epigenetic modifications to the pathogenesis of AD and makes recommendations for the treatment of AD by targeting epigenetic changes in the future.
Epigenetic modifications in the pathogenesis of AD
Effects of epigenetics on skin barrier
Epigenetic modifications in epidermal differentiation and formation
Epidermal lineages are derived from the ectoderm. During epidermal differentiation, the basal layer keratinocytes migrate upwards forming the spinous, granular and cornified differentiated layers. Normally structures and components of the epidermis maintain their integrities, including keratin, filaggrin (FLG), intercellular connection, keratinised envelope, lipid and calcium concentration, etc. Increasing evidence demonstrates that epigenetic modifications, including DNA methylation, histone modification and non-coding-RNAs, moderate epidermal formation and homeostasis.
Epigenetic programming and reprogramming seem to maintain pluripotency in early embryos and embryonic stem cells (ESCs) and DNA methylation confers epigenetic silencing of gene expression in ESCs.11 The major DNA methyltransferases (DNMTs) in mammals include DNMT1, DNMT3A and DNMT3B that establish and maintain DNA methylation patterns. DNMT1 is expressed in epidermal progenitor-containing cell populations to suppress differentiation and maintain basal cell proliferation.12 Many promoters of the differentiation-associated genes are hypermethylated and thus become suppressed during stratum corneum (SC) self-renewal.12 DNA methylation is also correlated with gene activation. DNMT3A/B was shown to bind to the active enhancers in an H3K36me3-dependent manner during human epidermal SCs differentiation.13 Specifically, DNMT3A associates with p63 to maintain high levels of DNA hydroxymethylation at the centre of enhancers in a Tet2-dependent manner.13 Whereas DNMT3B promotes DNA methylation along the body of the enhancer.13 In addition, DNMT3A/B is dispensable for murine epidermal development and homeostasis.14 Collectively, DNA methylation controls the proliferation and differentiation of the epidermis effectively [Table 1].
| Process | Epigenetic modification | Action mechanism | Reference |
|---|---|---|---|
| Epidermal differentiation | DNA methylation | DNMT1 suppressed differentiation and maintained basal cell proliferation | 12 |
| DNMT3A/B was bind to H3K36me3-dependent manner during SCs differentiation | 13 | ||
| PRC1-associated protein CBX4 maintained human epidermal balance | 15 | ||
| Histone proteins modifications | EZH2 methylation of H3K27me3 prevented premature recruitment of AP1 transcriptional activator | 18 | |
| Mll2 and its binding partner WDR5 regulated expression of differentiation-associated genes in human skin | 21 | ||
| 5AC and NaB promoted histone hyperacetylation to induce terminal differentiation through increasing expression of Sprr1/2 and involucrin in human keratinocytes | 23 | ||
| Epidermal dysfunction | DNA methylation | Excessive methylation of FLG may increase risk of AD | 44 |
| Histone proteins modifications | HDAC activation was associated with epithelial barrier dysfunction | 45 |
DNMT: DNA methyltransferase, SCs: stratum corneums, PRC1: polycomb repressive complex 1, 5AC: 5-azacytidine, NaB: sodium butyrate, FLG: filaggrin, AD: atopic dermatitis, HDAC: histone deacetylase
Mammalian polycomb group (PcG) proteins play central roles in maintaining stem cell pluripotency and regulating lineage-specific differentiation. Drosophila PcG proteins function within three major multimeric complexes: polycomb repressive complex 1 (PRC1), PRC2 and pleiohomeotic (Pho) repressive complex (PhoRC). In particular, a PRC1-associated protein CBX4 maintains human epidermal SCs in a slow-cycling and undifferentiated state.15 Luis et al. suggested that decreased keratinocyte proliferation and increased premature differentiation attribute to Cbx4 deletion in mouse epidermis.16 Moreover, mutant Cbx4 increased Cdkn2a/p16 transcripts which induced hyperproliferation and increased senescence in human keratinocytes.15 Also, transplantation of Cbx4-depleted keratinocytes failed to restore integral epithelium in mouse, indicating its significance in maintaining epidermal SCs.16 Three core subunits make up the catalytic core of PRC2—the SET domain containing EZH2, the zinc-finger containing SUZ12 and the WD40 repeat protein EED-which catalyses trimethylation on H3K27.17 EZH2 methylation of H3K27me3 prevented premature recruitment of AP1 transcriptional activator to the structural genes that are required for epidermal differentiation, and Ezh2 knock-out mouse showed premature differentiated skin layer in embryos.18 While basal cells differentiate, AP1 activates differentiation-specific genes as Ezh2 expression is lost.19 Altogether, these studies suggest that PRC components and H3K27me3 levels may influence skin SCs activation and maintenance. H3K4me3 is involved in gene activation which is mediated by the trithorax group (TrxG) component with a histone H3 lysine 4 (H3K4) methyltransferase activity.20 Particularly, the member Mll2 and its binding partner WDR5 are predominantly expressed in the differentiated cells and regulate the expression of differentiation-associated genes in human skin.21 Knockdown of Mll2 in human keratinocytes showed downregulation of differentiation-related genes as well.21 Therefore, H3K4 methylation is essential for keratinocyte-specific gene regulation. Moreover, Histone acetylation also participates in skin development. In vitro using HDAC inhibitor led foreskin tissue to abnormal epiderma structure, proliferation stagnate, and premature terminal differentiation.22 5-azacytidine (5AC) and sodium butyrate (NaB) promote histone hyperacetylation, which induces terminal differentiation through increased expression of Sprr1/2 and involucrin in human keratinocytes23 [Table 1].
Non-coding RNA (ncRNA) is a kind of RNA transcript without the function of coding protein, including microRNA, lncRNA, circRNA and so on. They are involved in many biological processes, such as cell proliferation, differentiation, apoptosis and immune response. Calcium gradients, transcriptional factors p63 and Notch are all involved in the regulation of epidermal keratinocyte differentiation and proliferation.24,25 Many studies reveal that multiple miRNAs are associated with this process via extracellular calcium. MiR-203, the first and the most upregulated miRNA implicated in epidermal differentiation, regulates calcium-induced keratinocyte differentiation by activation of the protein kinase C (PKC) and activator protein 1 (AP-1) pathway.26 Snail family transcriptional repressor 2 (SNAI2) and ΔNp63 are the targets of this process. MiR-203 inhibits p63 expression, which regulates keratinocyte proliferation and differentiation by galectin-7, resulting in upregulating the c-Jun N-terminal kinase (JNK).27 MiR-23b and the transforming growth factor-β (TGF-β)/SMAD signalling have a decisive function in regulating human epidermal differentiation.28,29 In addition, microRNA-23b-3p regulates human keratinocyte differentiation through repression of its direct target TGIF1 and activation of the TGF-β-SMAD2 signalling pathway.30 Extracellular Ca2+ increases miR-184 expression in primary epidermal keratinocytes and the upregulated miR-184 facilitates keratinocyte differentiation with increased involucrin expression via upregulation of cyclin E and p21 cyclin-dependent kinase inhibitors.31 Also, miR-184 promotes keratinocyte differentiation by enhancing the Notch pathway and targeting K15 and factor-inhibiting hypoxia-inducible factor 1 (FIH1).32 By contrast, p63 targets miRNAs via directly binding to miR-34a and miR-34c regulatory regions and repressing their activity.33 Moreover, overexpression of miR-34a could downregulate the sirtuin family member SIRT6 to stimulate keratinocyte differentiation34 [Table 2].
| Process | Epigenetic modification | Target molecule | Action mechanism | Reference |
|---|---|---|---|---|
| Epidermal differentiation | miR-203 | SNAI2 and ΔNp63 | Activate PKC and AP-1 pathway | 26 |
| P63 | Upregulated of JNK by galectin-7 | 27 | ||
| miR-23b-3p | TGIF1 | Interference in TGF-ß-SMAD2 signalling pathway | 30 | |
| miR-184 | Upregulated cyclin E and p21 cyclin-dependent kinase inhibitors | 31 | ||
| K15 and FIH1 | Enhanced the Notch pathway | 32 | ||
| miR-34a | Downregulated the sirtuin family member SIRT6 | 34 | ||
| miR-339-5p | Activation of the Wnt/β-catenin signalling pathway | 35 | ||
| miR-146a | EGFR | Inhibited keratinocyte proliferation | 36 | |
| H19 lncRNA | miR-130b-3p | Increased the expression of desmoglein 1 | 37 | |
| Epidermal dysfunction | miR-155-5p | PKIα | Inhibited expressions of tight junction proteins | 47 |
| miR-335 | Suppressed SOX6 expression | 48 | ||
| let-7a-5p | RRM2 and CCR7 | Barrier abnormalities | 49 | |
| miR-26a-5p | HAS3,DEPDC1B, DEPDC1,NAMPT, DENND1B and ADAM19 | 49 | ||
| miR-10a-5p | HAS3 | 50 |
SNAI2: snail family transcriptional repressor 2, PKC: protein kinase C, AP-1: activator protein 1, JNK: c-Jun N-terminal kinase, FIH1: factor-inhibiting hypoxia-inducible factor 1, EGFR: epidermal growth factor receptor, RRM2: ribonucleotide reductase regulatory subunit M2, CCR7: C-C motif chemokine receptor 7, HAS3: hyaluronan synthase 3, DEPDC: DEP domain-containing, NAMPT: nicotinamide phosphoribosyl transferase, DENND1B: DENN domain-containing 1B, ADAM19: a disintegrin and metalloproteinase domain 19
Downregulation of miR-339-5p increases levels of distal-less homeobox5 (DLX5), leading to involucrin upregulation via activation of the Wnt/β-catenin signalling pathway.35 Suppression of EGFR signalling facilitates terminal differentiation of keratinocytes. In addition, upregulation of miR-146a represses its target epidermal growth factor receptor (EGFR), thereby inhibiting keratinocyte proliferation.36 The interaction between H19 lncRNA and miR-130b-3p regulates desmoglein 1 expression. Specifically, H19 inhibits miR-130b-3p to increase the expression of desmoglein 1 to induce keratinocyte differentiation.37 Interestingly, the link between desmosomal cadherin and the stability of β-catenin, which is negatively regulated by miR-214 as a direct target, suggested the role of miR-214 in epidermal barrier function.38 In the context of calcium-induced keratinocyte differentiation, oleic acid, an unsaturated free fatty acid constituent of sebum, has been shown to accelerate keratinocyte differentiation via upregulation of miR-203.39 Linoleic acid and ciglitazone also increase sebaceous lipogenesis via upregulation of miR-203 and miR-574-3p in the differentiation process.40 Additionally, circRNA is abundantly expressed in the process of epidermal stem cell differentiation.41 Thus, we speculate that circRNAs may maintain skin integrity in AD which requires further investigation [Table 2].
Epigenetic modifications in the epidermal destruction and repair in AD
In the pathogenesis of AD, both FLG mutations and immune-mediated filaggrin deficiency could cause irregular or deformed KCs and damage the barrier.42,43 Studies by Ziyab et al. have shown that the excessive methylation of this gene may occur in heterozygotic carriers of null mutations (R501X, 2282del4 and S3247X); the interplay between the sequence and epigenetic regulation results in the increased risk of the disease in the carriers. These studies indicated that the epigenetic mechanisms regulating the expression of the FLG gene in the pathogenesis of the disease should not be overlooked.44 The expression of tight junctions such as transmembrane proteins (claudin; connecting adhesion molecules) and the intracellular cytoplasmic proteins (zonula clauddens, Zo) was decreased in AD. Steelant et al. described a new pathway of epithelial barrier dysfunction that was associated with histone deacetylase (HDAC) activation, and the application of HDAC inhibitors was effective to rebuild the barrier integrity and reduce hyperresponsiveness in asthmatic mice45 [Table 1].
Sonkoly et al. applied miRNA microarray to compare AD lesions with healthy controls and found miR-155 was one of the highest-ranked upregulated miRNAs in the patients’ group.46 Wang et al. also observed this result, they found that miR-155-5p predominantly increased in AD epithelium and PKIα was its specific target gene, through this pathway, the expressions of tight junction proteins (such as CLDN16, CLDND1 and occludin) were inhibited.47 MiR-335 directly suppressed SOX6 expression to induce keratinocytes differentiation. In AD lesions, miR-335 expression was abnormally decreased whereas SOX6 was overexpressed to induce epidermal dysfunction, and miR-335 was epigenetically regulated by HDACs as well.48 In addition, the downregulation of let-7a-5p may be related to barrier abnormalities by targeting ribonucleotide reductase regulatory subunit M2 (RRM2) and C-C motif chemokine receptor 7 (CCR7), which are mainly involved in cancer cell proliferation.49 Downregulated miR-26a-5p was also involved in the barrier function of patients with atopic dermatitis by targeting hyaluronan synthase 3 (HAS3), DEP domain-containing 1B (DEPDC1B), DEPDC1, nicotinamide phosphoribosyl transferase (NAMPT), DENN domain-containing 1B (DENND1B) and a disintegrin and metalloproteinase domain 19 (ADAM19), which mediated cell differentiation, cell proliferation and anti-apoptosis.49 HAS3 is one of the targets identified in downregulated miR-26a-5p.49 It is a direct target of upregulated miR-10a-5p in atopic dermatitis50 [Table 2].
Effects of epigenetics on immune dysfunction
Innate and adaptive immune systems are the main role players in the pathogenesis of AD. Innate immunity is the primary defensive barrier against pathogen infection, and adaptive immunity is critical in AD. Based on the adaptive inflammatory cascade reaction in AD, it has been described as a biphasic T cell disease. In the acute phase of AD, KCs are stimulated to release TSLP and other alarm proteins (such as IL-25 and IL-33). These proteins act on the ILCs to amplify Th2 response by inducing the transformation of immature T cells to Th2 cells, which promotes the production of cytokines that are related to Th2 (e.g., IL-4, IL-5, IL-13 and IL-31) and Th22 (e.g., IL-22 and S100A protein). The risk of developing allergy is influenced by early life events and particularly in utero exposures.51 In recent years, more and more articles reveal that epigenetic modifications also take place during pregnancy. Exposure to maternal smoking, BMI, DHA levels, vitamin D levels and folate supplementation have been shown to be associated with DNA methylation in cord blood.52 Hanna et al. demonstrated that maternal atopy is associated with specific epigenetic signatures in offspring. They found that some top CpG sites were mapped to genes SCD, ITM2C, NT5C3A and NPEPL1.52 Both ITM2C and NT5C3A were related to the immune system. ITM2C was a target gene of a T cell-specific transcription factor called GATA-3. ITM2C deficiency weakened T helper cell-dependent immune responses,53 which could influence the future development of Th2 phenotypes such as allergy. NT5C3A encoded a protein that affected erythrocyte function; its expression was induced by IFN-γ and acted as an anti-inflammatory regulator via an epigenetic mechanism.54 Children with low Treg levels at birth might possess a higher risk of developing AD or of sensitisation to food allergens in the first year of life.55 Human β-defensin-1 (HBD-1) is expressed in various epithelial tissues, including the skin. Methylation frequencies at the CpG 3 and 4 sites within the HBD-1 promoter were significantly higher in AD lesional samples, which led to abundant S. aureus colonisation.56 Epigenetic phenomena always regulate immune cell differentiation in the mechanism of AD. A region within the RAD50 gene on chromosome 5q31 controls Th2 differentiation, which is characterised by Rad50 DNase I hypersensitive sites (RHS). Polymorphism of rs2240032 in the RHS7 region affects the methylation of the Th2 cytokines promoter region and influences total serum IgE levels.57,58 Additionally, the overexpression of high-affinity IgE receptor (FCER1G) on monocytes and dendritic cells contributes to the pathogenesis of AD, as TSLP-activated pSTAT5 pathway regulates FCER1G demethylation on monocytes by recruiting DNA demethylase TET2.59 IFN-γ is strictly regulated during fetal development, its gene transcription has a relevance to the methylation status of a CpG dinucleotide contained within a TATA proximal regulatory element.60 A study nested in the prospective birth cohort ALADDIN (Assessment of Infant Lifestyle and Allergic Diseases) showed that placental histone hyperacetylation was associated with a low risk of food allergen sensitisation in children.61 They observed H3 acetylation in IFNG gene, H4 acetylation in HDAC4 and H3 acetylation in SH2B3.61 H4 acetylation increased HDAC4 expression in the placenta; immune genes and transcription factors expression involved in Th1 tilting may be regulated by whole-genome deacetylase to improve early immune polarisation, which may help infants to prevent IgE sensitisation.61 Admittedly, ncRNAs also regulate immune function in AD. Th17 cell differentiation is controlled by retinoid-related orphan receptor gamma t (RORγt). DEAD-box protein 5 (DDX5) is identified as a co-activator of RORγt, which is necessary for the transcription of Th17 genes involved in Th17-mediated autoimmune inflammation. Interestingly, lncRNA RMRP contributes to forming the DDX5-RORγt complex and recruiting this complex to DNA binding sites in Th17 cells.62 In addition, a study mentioned that miR-155, FOXP3 and RORγt may have indefinite relevance to each other to mediate immune disorders in AD.63 Tregs are best characterised by the expression of forkhead box transcription factor 3 (Foxp3), whose stable expression during Treg differentiation is regulated by several conserved non-coding sequence (CNS) elements. Owing to the unique catalysis by methylcytosine dioxygenase of the ten-eleven-translocation (Tet) family, CNS2 is completely demethylated to maintain Treg cell suppressor function.64 Foxp3-TSDR hypermethylation of Tregs was observed in both allergic rhinitis and AD.55,65 The euchromatin histone methyltransferase-1 (Ehmt1) and Ehmt2 play a considerable role in the epigenetic events of euchromatin. They preferentially establish heterodimerisation to synthesise the critical methyltransferase and catalyse the monomethylation and demethylation of lysine on histone (h3k9me1/me2).66 Treg cell differentiation relies on the epigenetic state of Foxp3-TSDR, within which the Ehmt1 activity is a fundamental control mechanism. The transcription factor Wiz recruited Ehmt1 to Foxp3-TSDR elements to make a preference for methylation. Knocking out of Ehmt1 or Wiz by CRISPR/Cas method led to the loss of H3K9me2 and enhancement of Foxp3 expression during iTreg differentiation67 [Figure 1 and Table 3].
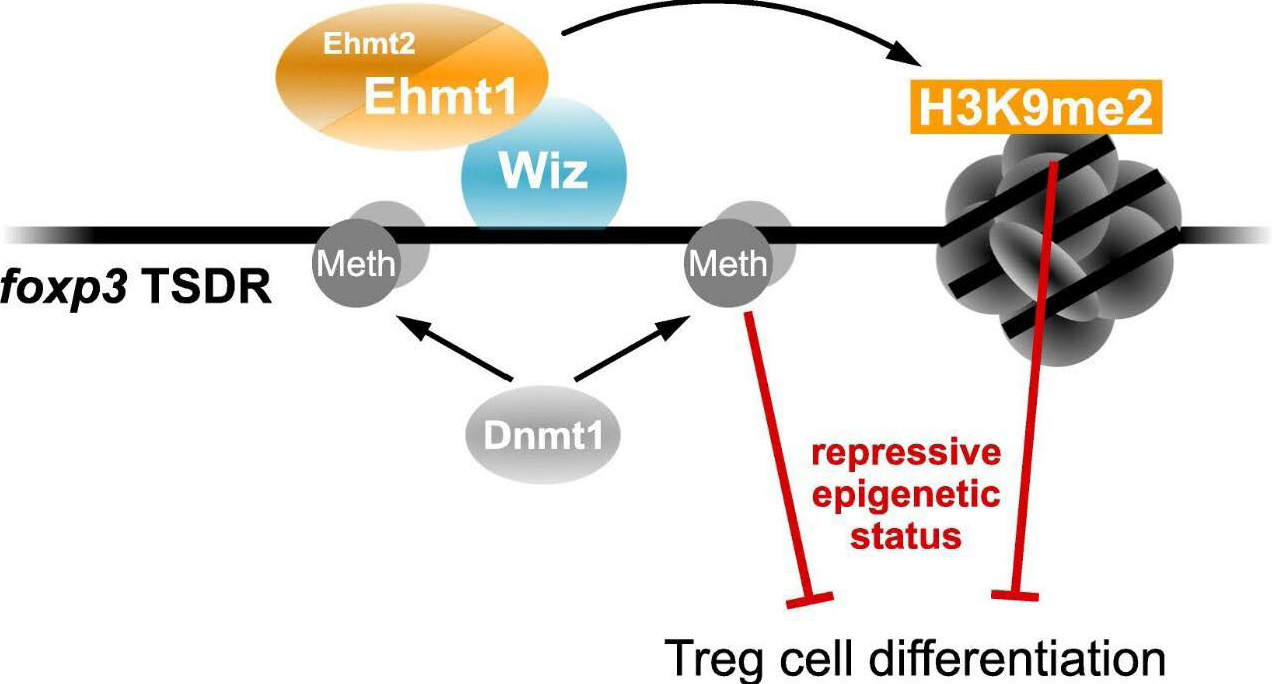
- Wiz recruited Ehmt1 to Foxp3-TSDR elements to induce Tregs differentiation
| Process | Epigenetic modification | Action mechanism | Reference |
|---|---|---|---|
| Immune dysfunction | DNA methylation | ITM2C deficiency weakened T helper cell-dependent immune responses | 53 |
| NT5C3A expression was induced by IFN-γ and acted as an anti-inflammatory regulator via an epigenetic mechanism | 54 | ||
| HBD-1 methylation in AD lesional samples led to abundant S. aureus colonisation | 56 | ||
| RHS7 region affected the methylation of the Th2 cytokines promoter region and influenced total serum IgE levels | 57,58 | ||
| Histone proteins modifications | H4 acetylation in HDAC4 helped infants to prevent IgE sensitisation | 61 |
HBD-1: human β-defensin-1, AD: atopic dermatitis, RHS7: Rad50 DNase I hypersensitive sites, HDAC4: histone deacetylase 4
Effects of epigenetics on S. aureus colonisation
Research shows a high prevalence S. aureus colonisation in lesions of AD patients, and its abundance positively correlates with disease severity.68 The mechanisms of S. aureus colonisation in AD include enhancing skin adhesion, damaging epidermal barrier function and amplifying proinflammatory reactions. S. aureus produces various cell-wall proteins and secreted toxins that make it adhere to the human skin surface and destroy the skin barrier through physical, chemical and inflammatory mechanisms. Several adhesive molecules such as fibronectin-binding protein (fnBP), clumping factors A and B (ClfA and ClfB) and iron-regulated surface determinant A (IsdA) have been found. S. aureus alpha-toxin, an effective pore-forming cytotoxin, forms a heptameric beta-barrel pore in the cell membranes and corrodes epidermal integrity.69 Moreover, S. aureus produces a variety of proteases to accelerate SC dissolution, and the activity of these proteases would be enhanced when filaggrin decreases and typical Th2 cytokines exist.70 Also, S. aureus could irritate endogenous keratinocyte proteases (e.g., KLK6, KLK13 and KLK14) to create a mileu favouring epidermal destruction.71 In addition, S. aureus colonisation induces a series of proinflammatory cytokine production. Staphylococcal superantigens (SAgs), such as SEA, SEB, SEC and toxic shock syndrome toxin-1 (TSST-1), drive B-cell development and cytokine secretion. SAgs also induce the release of IL-31, which inhibits keratinocyte differentiation, down-regulates filaggrin expression and stimulates itch.72 Remarkably, Staphylococcal lipoproteins facilitated the expression of TSLP in keratinocytes through the toll-like receptor 2/6 (TLR-2/6) pathway to strengthen Th2 responses.73 S. aureus secretes several short amphiphilic peptides called phenol-soluble modulins (PSMs) which are directly acting pro-inflammatory factors with compartment-specific effects. PSMs initiate γδT-mediated inflammation driven by IL-36a in the epidermal compartment, whereas in the dermal compartment, they stimulate IL-1b-driven TH17 inflammation.74,75 In conclusion, S. aureus colonisation plays a critical role in the pathogenesis of AD. One study proposed that S. aureus infection probably induced histone post-translational modification, which led to an abnormal expression of important signalling pathways participating in the inflammatory response, immune monitoring and bacterial elimination.76 However, the available literature is limited; epigenetic changes caused by S. aureus colonisation are in need of further investigation.
Epigenetics in skin and intestinal dysbiosis
Variations in cutaneous and intestinal microbiota contribute to the pathogenesis of AD. The “gut-immune-skin axis” considers that both skin and intestinal microecosystems have a significant influence on developing the immune system and forming immune tolerance in early life, and these microecosystems may affect each other through immune responses. For infants, the activation of TLRs increases IL-6 and IL-23 levels and decreases tumour necrosis factor (TNF) -α or IL-1 levels. This reaction facilitated the body’s immune tolerance to self or foreign antigens and weakened the skin response to microorganisms, which was relatively obvious for older children and adults. Furthermore, several critical microbes enhanced inflammatory reactions. For instance, the epidermal T cells produced IL-17 A and IFN-γ after Staphylococcus epidermidis antigen stimulation.77 In particular, the immune responses of Th17 cells could be extensively triggered by skin-colonising microbiota.78
On the other hand, abundant evidence supports the role of intestinal microbes in the pathogenesis of AD mediated by immunologic, metabolic and neuroendocrine pathways. The specific composition of early intestinal microbiomes regulates the host’s immune maturation. Although S. aureus colonisation in the skin is positively related to the severity of AD, a birth cohort study showed that AD was negatively correlated with intestinal colonisation of S. aureus strains carrying superantigens and adhesion genes into infancy. So it is speculated that intestinal colonisation by this particular bacterium provides protection by promoting immune system development.79 Feedback interactions between Faecalibacterium prausnitzii imbalance and intestinal epithelial inflammation lead to increasing gut epithelial permeability and abnormal Th2-type immune responses in the skin ultimately.80 Additionally, the intestinal microbiome affects the immune system through its metabolites as well. A higher proportion of gut microbiota in AD patients produces short-chain fatty acids (SCFAs), whose products include butyrate, propionate and acetate, all of which interact with the gut epithelium barrier and execute anti-inflammatory and immune-modulatory effects.80 Moreover, supplementation of linoleic acid and 10-hydroxy-cis-12-octadecenoic acid could alleviate AD symptoms and help recover the gut microecosystem balance.81 Interestingly, multiple neurotransmitters and neuromodulators are also involved in the intestinal microbiological composition, which includes tryptophan, γ-aminobutyric acid and serotonin, all of which are associated with AD severity, skin barrier disturbance and immune system disorders.82,83
Therefore, both the skin and intestinal microbiome experience a non-negligible variation in AD patients. The diversity of microbiota is decreased, and the amount of Streptococcus, Corynebacterium, Keratinella and Proteobacteria are generally reduced, while the abundance of S. aureus in the skin is prominently increased.84 The microbial diversity of the skin is restored after treatment. Compared with healthy persons, AD patients were observed to have higher levels of Clostridia, Clostridium difficile, Escherichia coli and S. aureus and lower levels of Bifidobacteria, Bacteroidetes and Bacteroides in the gut.85 In particular, the abundance of butyric-producing bacteria (e.g., Coprococcuseutactus) in slight-mild AD or healthy infants was higher than that in severe AD. It remains uncertain, however, whether or not abnormal changes in skin and intestinal microbes are the primary results of the destruction of the epidermal barrier and the bias of Th2-type immune responses. Although research on skin and intestinal microecology variation in AD has been gradually expanding, there is still a little acquaintance with the epigenetic changes caused by microecology. Butyric acid is a metabolite of SCFs in S. epidermidis. BA-NH-NH-BA, a derivative of butyric acid, could induce the acetylation at lysine 9 of H3 (AcH3K9) in keratinocytes to serve as an HDAC inhibitor and could increase IL-6 production to reduce S. aureus colonisation prominently.86 Conversely, environmental factors may damage the epidermal barrier, cause immune disorders and participate in proinflammatory responses induced by S.aureus infection through histone modification, which has become established a non-negligible mechanism of AD. Interestingly, Ansari et al. discovered that the intestinal epithelium exposed to commensal microbiota induced localised TET2/3-dependent DNA methylation changes at regulatory elements, and this microbiota-induced epigenetic programming was necessary for proper intestinal homeostasis in vivo. 87 The chances are that we can focus on the mechanism of this process thoroughly in the future.
Others
There are also some other mechanisms in AD, including vitamin D deficiency, histamine H4 receptor function and cell death. Research indicates that the average level of 1,25-(OH)2-VitD3 in AD patients is significantly reduced, and the severity of the disease is dose-dependent.88 Furthermore, 1-hydroxylase and 2,4-hydroxylase of vitamin D are under epigenetic control.89,90 Distinct methylation levels of CYP2R1 and CYP24A1 were detected between vitamin D deficient groups and sufficient controls.91 Histamine H4 receptor(H4R) is the latest hotspot in the histamine receptor family and its antagonist was able to alleviate patients’ eczematoid lesions, itch and inflammatory reactions, which provide a creative therapeutic target of AD.92,93 MiR-223 was increased in AD, it may up-regulate histamine-N-methyltransferase (HNMT) expression indirectly to degrade the excessive histamine involved in the pathogenesis of AD.94 Keratinocytes in AD patients showed increased IFN-γ-mediated apoptosis, and apoptosis-related genes NOD2, DUSP1 and ADM were all induced by the IFN-γ program.95 Remarkably, miR-29b was significantly upregulated and directly targeted the BCL2L2 gene to promote IFN-γ-induced keratinocyte apoptosis.96 So, all of the above processes are regulated by epigenetics in AD [Table 4].
| Process | Epigenetic modification | Target molecule | Action mechanism | Reference |
|---|---|---|---|---|
| Others | MiR-223 | Up-regulated HNMT to degrade the excessive histamine | 94 | |
| miR-29b | BCL2L2 | Promote IFN-γ-induced keratinocyte apoptosis | 96 |
HNMT: histamine-N-methyltransferase
Conclusion
Pathogenetically, AD has multiple pathways of clinical evolution, including the destruction of the epidermal barrier, activation of multifarious T cell subsets and commensal bacteria dysbiosis. Indeed, it has been well proven that environmental factors and diets mediate the pathogenesis of allergic diseases, such as asthma, allergic rhinitis, AD and food allergy through epigenetic mechanisms. In the current treatments of AD, drugs such as antihistamines, corticosteroids, leukotriene modifiers, anti-IgE agents, anticholinergics and beta-agonists have been proven capable of effectively controlling allergic symptoms, but AD recurrence is still a worldwide problem.97 Fortunately, several methods targeting epigenetic modifications have been therapeutically validated in treating AD. For instance, anti-miR-155-5p and anti-miR-126 down-regulated inflammatory cytokines and inhibited airway hyperresponsiveness in airway allergic diseases.98,99 Liew et al. demonstrated that belinostat, the inhibitor of histone deacetylase, restored miR-335 expression and rescued the defective skin barrier in AD by targeting the dysregulated miR-335/SOX6 axis.48 Trichostatin A (TSA) prevented CD4+ T cells from producing IL-4 cytokine to alleviate symptoms in AD mouse models.100 And JNJ-26481585 restored nasal mucosa function by promoting tight junction components expression.45 However, a thorough understanding of the epigenetic modifications may provide the base for new molecular classifications of the disease and the development of personalised therapies, which requires our further efforts.
Declaration of patient consent heading
Patient’s consent not required as patient’s identity is not disclosed or compromised.
Financial support and sponsorship
This study was supported by the Fundamental Research Funds for the Central Universities of Central South University (No. 1053320192081).
Conflicts of interest
There are no conflicts of interest.
References
- Long-term Western diet intake leads to dysregulated bile acid signaling and dermatitis with Th2 and Th17 pathway features in mice. J Dermatol Sci. 2019;95:13-20.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- A short period of breastfeeding in infancy, excessive house cleaning, absence of older sibling, and passive smoking are related to more severe atopic dermatitis in children. Eur J Dermatol. 2018;28:56-63.
- [CrossRef] [PubMed] [Google Scholar]
- What is the evidence for interactions between filaggrin null mutations and environmental exposures in the aetiology of atopic dermatitis? A systematic review. Br J Dermatol. 2020;183:443-51.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Atopic dermatitis is associated with active and passive cigarette smoking in adolescents. PLoS One. 2017;12:e0187453.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- The interaction between epigenetics, nutrition and the development of cancer. Nutrients. 2015;7:922-47.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Epigenetics and allergy: From basic mechanisms to clinical applications. Epigenomics. 2017;9:539-71.
- [CrossRef] [PubMed] [Google Scholar]
- Atopic dermatitis: Interaction between genetic variants of GSTP1, TNF, TLR2, and TLR4 and air pollution in early life. . 2018;29:596-605.
- [CrossRef] [PubMed] [Google Scholar]
- MicroRNA-143 inhibits IL-13-induced dysregulation of the epidermal barrier-related proteins in skin keratinocytes via targeting to IL-13Ralpha1. Mol Cell Biochem. 2016;416:63-70.
- [CrossRef] [PubMed] [Google Scholar]
- Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425-32.
- [CrossRef] [PubMed] [Google Scholar]
- DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature. 2010;463:563-7.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Dnmt3a and Dnmt3b associate with enhancers to regulate human epidermal stem cell homeostasis. Cell Stem Cell. 2016;19:491-501.
- [CrossRef] [PubMed] [Google Scholar]
- Loss of Dnmt3a and Dnmt3b does not affect epidermal homeostasis but promotes squamous transformation through PPAR-γ. eLife. 2017;6:e21697.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Cbx4 maintains the epithelial lineage identity and cell proliferation in the developing stratified epithelium. J Cell Biol. 2016;212:77-89.
- [Google Scholar]
- Regulation of human epidermal stem cell proliferation and senescence requires polycomb-dependent and -independent functions of Cbx4. Cell Stem Cell. 2011;9:233-46.
- [CrossRef] [PubMed] [Google Scholar]
- Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat Commun. 2016;7:11316.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122-35.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Epigenetic regulation of skin: Focus on the Polycomb complex. Cellular and molecular life sciences. CMLS. 2012;69:2161-72.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Polycomb/trithorax antagonism: Cellular memory in stem cell fate and function. Cell Stem Cell. 2019;24:518-33.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- GRHL3/GET1 and trithorax group members collaborate to activate the epidermal progenitor differentiation program. PLoS Genet. 2012;8:e1002829.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Inhibition of histone deacetylation promotes abnormal epidermal differentiation and specifically suppresses the expression of the late differentiation marker profilaggrin. J Invest Dermatol. 2007;127:1126-39.
- [CrossRef] [PubMed] [Google Scholar]
- Evidence for local control of gene expression in the epidermal differentiation complex. Exp Dermatol. 2002;11:406-12.
- [CrossRef] [PubMed] [Google Scholar]
- Calcium—A central regulator of keratinocyte differentiation in health and disease. Eur J Dermatol. 2014;24:650-61.
- [CrossRef] [PubMed] [Google Scholar]
- Transcriptional control of late differentiation in human keratinocytes by TAp63 and Notch. Exp Dermatol. 2015;24:754-60.
- [CrossRef] [PubMed] [Google Scholar]
- Protein kinase C-dependent upregulation of miR-203 induces the differentiation of human keratinocytes. J Invest Dermatol. 2010;130:124-34.
- [CrossRef] [PubMed] [Google Scholar]
- Galectin-7 regulates keratinocyte proliferation and differentiation through JNK-miR-203-p63 signaling. J Invest Dermatol. 2016;136:182-91.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- A comprehensive analysis of microRNA expression during human keratinocyte differentiation in vitro and in vivo. J Invest Dermatol. 2011;131:20-9.
- [CrossRef] [PubMed] [Google Scholar]
- A decisive function of transforming growth factor-β/Smad signaling in tissue morphogenesis and differentiation of human HaCaT keratinocytes. Mol Biol Cell. 2011;22:782-94.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- MicroRNA-23b-3p regulates human keratinocyte differentiation through repression of TGIF1 and activation of the TGF-ß-SMAD2 signalling pathway. Exp Dermatol. 2017;26:51-7.
- [CrossRef] [PubMed] [Google Scholar]
- microRNA-184 is induced by store-operated calcium entry and regulates early keratinocyte differentiation. J Cell Physiol. 2020;235:6854-61.
- [CrossRef] [PubMed] [Google Scholar]
- microRNA-184 induces a commitment switch to epidermal differentiation. Stem Cell Rep. 2017;9:1991-2004.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Transcriptional repression of miR-34 family contributes to p63-mediated cell cycle progression in epidermal cells. J Investig Dermatol. 2010;130:1249-57.
- [CrossRef] [PubMed] [Google Scholar]
- Differential microRNA expression profile comparison between epidermal stem cells and differentiated keratinocytes. Mol Med Rep. 2015;11:2285-91.
- [CrossRef] [PubMed] [Google Scholar]
- miR-339-5p negatively regulates loureirin A-induced hair follicle stem cell differentiation by targeting DLX5. Mol Med Rep. 2018;18:1279-86.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Down-regulation of microRNA-146a is associated with high-risk human papillomavirus infection and epidermal growth factor receptor overexpression in penile squamous cell carcinoma. Hum Pathol. 2017;61:33-40.
- [CrossRef] [PubMed] [Google Scholar]
- H19 lncRNA regulates keratinocyte differentiation by targeting miR-130b-3p. Cell Death Dis. 2017;8:e3174.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- MicroRNA-214 controls skin and hair follicle development by modulating the activity of the Wnt pathway. J Cell Biol. 2014;207:549-67.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Oleic acid enhances keratinocytes differentiation via the upregulation of miR-203 in human epidermal keratinocytes. J Cosmet Dermatol. 2019;18:383-9.
- [CrossRef] [PubMed] [Google Scholar]
- Differentially regulated microRNAs during human sebaceous lipogenesis. J Dermatol Sci. 2013;70:88-93.
- [CrossRef] [PubMed] [Google Scholar]
- Circular RNAs are abundantly expressed and upregulated during human epidermal stem cell differentiation. RNA Biol. 2018;15:280-91.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- The IL-13-OVOL1-FLG axis in atopic dermatitis. Immunology. 2019;158:281-6.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- A common variant on chromosome 11q13 is associated with atopic dermatitis. Nat Genetics. 2009;41:596-601.
- [CrossRef] [PubMed] [Google Scholar]
- DNA methylation of the filaggrin gene adds to the risk of eczema associated with loss-of-function variants. J Eur Acad Dermatol Venereol. 2013;27:e420-3.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Blocking histone deacetylase activity as a novel target for epithelial barrier defects in patients with allergic rhinitis. J Allergy Clin Immunol. 2019;144:1242-53.e7.
- [CrossRef] [PubMed] [Google Scholar]
- MiR-155 is overexpressed in patients with atopic dermatitis and modulates T-cell proliferative responses by targeting cytotoxic T lymphocyte-associated antigen 4. J Allergy Clin Immunol. 2010;126:581-9.e1-20.
- [CrossRef] [PubMed] [Google Scholar]
- Lower vitamin D levels in the breast milk is associated with atopic dermatitis in early infancy. Pediatr Allergy Immunol. 2019;31:258-64.
- [CrossRef] [PubMed] [Google Scholar]
- Belinostat resolves skin barrier defects in atopic dermatitis by targeting the dysregulated miR-335:SOX6 axis. J Allergy Clin Immunol. 2020;146:606-620.e12.
- [CrossRef] [PubMed] [Google Scholar]
- Identification and interaction analysis of key genes and microRNAs in atopic dermatitis by bioinformatics analysis. Clin Exp Dermatol. 2019;44:257-64.
- [CrossRef] [PubMed] [Google Scholar]
- MicroRNAs in atopic dermatitis: A systematic review. J Cell Mol Med. 2020;24:5966-72.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- In utero programming of allergic susceptibility. Int Arch Allergy Immunol. 2016;169:80-92.
- [CrossRef] [PubMed] [Google Scholar]
- Maternal atopy and offspring epigenome-wide methylation signature. Epigenetics. 2021;16:629-41.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Itm2a, a target gene of GATA-3, plays a minimal role in regulating the development and function of T cells. PLoS One. 2014;9:e96535.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- The intracellular pyrimidine 5’-nucleotidase NT5C3A is a negative epigenetic factor in interferon and cytokine signaling. Science Signal. 2018;11:eaal2434.
- [CrossRef] [PubMed] [Google Scholar]
- Cord blood Tregs with stable FOXP3 expression are influenced by prenatal environment and associated with atopic dermatitis at the age of one year. Allergy. 2012;67:380-9.
- [CrossRef] [PubMed] [Google Scholar]
- Promoter DNA methylation contributes to human beta-defensin-1 deficiency in atopic dermatitis. Animal Cells Syst. 2018;22:172-7.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- A polymorphism in the TH 2 locus control region is associated with changes in DNA methylation and gene expression. Allergy. 2014;69:1171-80.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- A common atopy-associated variant in the Th2 cytokine locus control region impacts transcriptional regulation and alters SMAD3 and SP1 binding. Allergy. 2014;69:632-42.
- [CrossRef] [PubMed] [Google Scholar]
- Thymic stromal lymphopoietin epigenetically upregulates Fc receptor gamma subunit-related receptors on antigen-presenting cells and induces TH2/TH17 polarization through dectin-2. J Allergy Clin Immunol. 2019;144:1025-35.e7.
- [CrossRef] [PubMed] [Google Scholar]
- Differentiation of the T helper phenotypes by analysis of the methylation state of the IFN-gamma gene. J Immunol. 1994;153:3603-10.
- [PubMed] [Google Scholar]
- Epigenetic modifications in placenta are associated with the child’s sensitization to allergens. BioMed Res Int. 2019;2019:1315257.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Retraction Note: DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature. 2018;562:150.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Expression of miRNA 155, FOXP3 and ROR gamma, in children with moderate and severe atopic dermatitis. G Ital Dermatol Venereol. 2017;155:168-72.
- [CrossRef] [PubMed] [Google Scholar]
- DNA demethylation of the Foxp3 enhancer is maintained through modulation of ten-eleven-translocation and DNA methyltransferases. Mol Cells. 2016;39:888-97.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Down-regulation of Tet2 is associated with Foxp3 TSDR hypermethylation in regulatory T cell of allergic rhinitis. Life Sci. 2020;241:117101.
- [CrossRef] [PubMed] [Google Scholar]
- Methylation of DNA ligase 1 by G9a/GLP recruits UHRF1 to replicating DNA and regulates DNA methylation. Mol Cell.. 2017;67:550-65.e5.
- [CrossRef] [PubMed] [Google Scholar]
- Recruitment of histone methyltransferase Ehmt1 to Foxp3 TSDR counteracts differentiation of induced regulatory T cells. J Mol Biol. 2019;431:3606-25.
- [CrossRef] [PubMed] [Google Scholar]
- Staphylococcus aureus density on lesional and nonlesional skin is strongly associated with disease severity in atopic dermatitis. J Allergy Clin Immunol. 2016;137:1272-4.e3.
- [CrossRef] [PubMed] [Google Scholar]
- Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859-66.
- [CrossRef] [PubMed] [Google Scholar]
- Staphylococcus aureus exploits epidermal barrier defects in atopic dermatitis to trigger cytokine expression. J Investig Dermatol. 2016;136:2192-200.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Staphylococcus aureus: Master manipulator of the skin. Cell Host Microbe. 2017;22:579-81.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- IL-31 regulates differentiation and filaggrin expression in human organotypic skin models. J Allergy Clin Immunol. 2012;129:426-33, 433.e1–8.
- [CrossRef] [PubMed] [Google Scholar]
- Staphylococcus aureus membrane and diacylated lipopeptide induce thymic stromal lymphopoietin in keratinocytes through the Toll-like receptor 2-Toll-like receptor 6 pathway. J Allergy Clin Immunol. 2010;126:985-93, 993.e1–3.
- [CrossRef] [PubMed] [Google Scholar]
- Staphylococcus aureus epicutaneous exposure drives skin inflammation via IL-36-mediated T cell responses. Cell Host Microbe. 2017;22:653-666.e5.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Staphylococcus aureus virulent PSMalpha peptides induce keratinocyte alarmin release to orchestrate IL-17-dependent skin inflammation. Cell Host Microbe. 2017;22:667-677.e5.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Epigenetic response in mice mastitis: Role of histone H3 acetylation and microRNA(s) in the regulation of host inflammatory gene expression during Staphylococcus aureus infection. Clin Epigenet. 2014;6:12.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Human TH9 cells are skin-tropic and have autocrine and paracrine proinflammatory capacity. Science Transl Med. 2014;6:219ra8.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature. 2015;520:104-8.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Superantigens and adhesins of infant gut commensal Staphylococcus aureus strains and association with subsequent development of atopic eczema. Br J Dermatol. 2017;176:439-45.
- [CrossRef] [PubMed] [Google Scholar]
- Faecalibacterium prausnitzii subspecies-level dysbiosis in the human gut microbiome underlying atopic dermatitis. J Allergy Clin Immunol. 2016;137:852-60.
- [CrossRef] [PubMed] [Google Scholar]
- Supplemental feeding of a gut microbial metabolite of linoleic acid, 10-hydroxy-cis-12-octadecenoic acid, alleviates spontaneous atopic dermatitis and modulates intestinal microbiota in NC/nga mice. Int J Food Sci Nutr. 2017;68:941-51.
- [CrossRef] [PubMed] [Google Scholar]
- Mind-altering microorganisms: The impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13:701-12.
- [CrossRef] [PubMed] [Google Scholar]
- Microbiome-derived tryptophan metabolites and their aryl hydrocarbon receptor-dependent agonist and antagonist activities. Mol Pharmacol. 2014;85:777-88.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Sci Transl Med. 2017;9:eaal4651.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Microbiosis in pathogenesis and intervention of atopic dermatitis. Int Immunopharmacol. 2019;69:263-69.
- [CrossRef] [PubMed] [Google Scholar]
- A derivative of butyric acid, the fermentation metabolite of Staphylococcus epidermidis, inhibits the growth of a Staphylococcus aureus strain isolated from atopic dermatitis patients. Toxins. 2019;11:311.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- The microbiota programs DNA methylation to control intestinal homeostasis and inflammation. Nat Microbiol. 2020;5:610-9.
- [CrossRef] [PubMed] [Google Scholar]
- Dose-response association between vitamin D deficiency and atopic dermatitis in children, and effect modification by gender: A case-control study. J Dermatol Treat. 2021;32:174-9.
- [CrossRef] [PubMed] [Google Scholar]
- Epigenetic mechanisms in multiple sclerosis and the major histocompatibility complex (MHC) Discov Med. 2011;11:187-96.
- [PubMed] [Google Scholar]
- Placenta-specific methylation of the vitamin D 24-hydroxylase gene: Implications for feedback autoregulation of active vitamin D levels at the fetomaternal interface. J Biol Chem. 2009;284:14838-48.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- A genome-wide methylation study of severe vitamin D deficiency in African American adolescents. J Pediatr. 2013;162:1004-9.e1.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Phase 2a, randomized, double-blind, placebo-controlled, multicenter, parallel-group study of a H4 R-antagonist (JNJ-39758979) in Japanese adults with moderate atopic dermatitis. J Dermatol. 2015;42:129-39.
- [CrossRef] [PubMed] [Google Scholar]
- Efficacy and safety of the histamine H4 receptor antagonist ZPL-3893787 in patients with atopic dermatitis. J Allergy Clin Immunol. 2019;143:1830-37.e4.
- [CrossRef] [PubMed] [Google Scholar]
- MicroRNA-223 is involved in the pathogenesis of atopic dermatitis by affecting histamine-N-methyltransferase. Cell Mol Biol. 2018;64:103-7.
- [CrossRef] [PubMed] [Google Scholar]
- Mechanisms of IFN-gamma-induced apoptosis of human skin keratinocytes in patients with atopic dermatitis. J Allergy Clin Immunol. 2012;129:1297-306.
- [CrossRef] [PubMed] [Google Scholar]
- IFN-gamma-induced microRNA-29b up-regulation contributes to keratinocyte apoptosis in atopic dermatitis through inhibiting Bcl2L2. Int J Clin Exp Pathol. 2017;10:10117-26.
- [PubMed] [PubMed Central] [Google Scholar]
- Specific allergen immunotherapy for the treatment of atopic eczema: A Cochrane systematic review. Allergy. 2016;71:1345-56.
- [CrossRef] [PubMed] [Google Scholar]
- MicroRNA expression is altered in an ovalbumin-induced asthma model and targeting miR-155 with antagomirs reveals cellular specificity. PLoS One. 2015;10:e0144810.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Altered expression of microRNA in the airway wall in chronic asthma: MiR-126 as a potential therapeutic target. BMC Pulm Med. 2011;11:29.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- The role of epigenetics in allergy and asthma development. Curr Opin Allergy Clin Immunol. 2020;20:48-55.
- [CrossRef] [PubMed] [Google Scholar]






