Translate this page into:
Filaggrin gene polymorphisms in Indian children with atopic dermatitis: A cross-sectional multicentre study
Corresponding author: Dr. Sahana M Srinivas, Department of Pediatric Dermatology, Indira Gandhi Institute of Child Health, Bengaluru, Karnataka, India. sahana.bmc@gmail.com
-
Received: ,
Accepted: ,
How to cite this article: Srinivas SM, Dhar S, Gowdra A, Saha A, Sundararajan L, Geetha TS, et al. Filaggrin gene polymorphisms in Indian children with atopic dermatitis: A cross-sectional multicentre study. Indian J Dermatol Venereol Leprol 2023;89:819-27.
Abstract
Background
Filaggrin (FLG) gene encoding the protein filaggrin plays an important role in barrier function of the skin and its alteration is a predisposing factor for atopic dermatitis. FLG gene variants result in absent or decreased filaggrin protein. Worldwide, the prevalence of FLG variants ranges from 14 to 56%. FLG null variants are distinct in each population.
Objectives
To study the FLG gene polymorphisms in Indian children and attempt a genotype-phenotype correlation in atopic dermatitis.
Methods
This was a cross-sectional, multicentre study conducted on 75 Indian children. Demographic details, clinical features and identified FLG null variants were recorded. We performed a whole gene sequencing of the entire FLG coding region using next-generation sequencing technology.
Results
The prevalence of FLG null variants was 34.7%. A total of 20 different FLG loss of function variants in 26 children were documented. Sixteen (80%) variants were novel and four (20%) were previously reported in Asian and European populations. We found a statistically significant association between FLG variants with early age of onset of atopic dermatitis (P = 0.016) and elevated serum IgE levels (P = 0.051). There was no significant difference between atopic dermatitis phenotypes in children having one variant as compared to children harbouring two or more null variants.
Limitation
Small sample size.
Conclusion
Our study reports a unique set of FLG variants different from Asian and European populations, with these variants being significantly associated with an early age of onset of atopic dermatitis and elevated serum IgE levels.
Keywords
Atopic dermatitis
early age
filaggrin polymorphisms
next-generation sequencing
null variants
serum IgE
Plain Language Summary
Atopic dermatitis is a chronic, itchy, relapsing inflammatory skin disorder seen more commonly in children. Filaggrin is a protein which plays an important role in maintaining the normal barrier function of the skin. Defects in the filaggrin gene can cause reduced or absent production of filaggrin, contributing to the development of atopic dermatitis. This study was conducted on 75 children at two different hospitals in India, to identify the filaggrin gene variants and assess their role in atopic dermatitis. There were 20 different filaggrin gene variants identified in 26 children in our study. Among them, 16 filaggrin gene variants were unique to our study population while four had been reported previously in Asian and European populations. These filaggrin gene variants in our study were significantly associated with an early age of onset of the atopic dermatitis and increased serum levels of the IgE class of antibodies.
Introduction
Atopic dermatitis is a chronic, progressive, relapsing inflammatory skin disorder characterised by varying degrees of pruritus and inflammation seen in 20% of children and 1–3% of adults.1 The prevalence of atopic dermatitis in India is around 0.5–7.2 per cent.2 Atopic dermatitis is the result of a complex interplay of several environmental, dietary, behavioural and immunological factors in genetically predisposed individuals.2 Filaggrin (filament aggregating protein, FLG) gene is an important component of skin barrier function and plays an important role in epidermal differentiation. Epidermal barrier and FLG null variants are thought to play a key role in the pathogenesis of atopic dermatitis, as well as in allergic rhinitis and asthma.3 Null variation is a change in the gene that results in non-transcription of mRNA or translation of protein or both. Null variants can be due to frameshifts or nonsense substitutions. FLG gene studies have been carried out in many geographical populations, which include European and Asian populations (Japanese, Chinese, Korean, Taiwan and Singaporean). Patients with FLG variants have been reported to have more persistent and severe disease, a higher incidence of herpes virus infections, association with ichthyosis vulgaris, palmar hyperlinearity, keratosis pilaris, asthma and a greater risk of multiple allergies than patients with atopic dermatitis without FLG variants.4 FLG gene variants may predict early onset, severe disease, worse prognosis, persistence of the disease in adulthood, allergen sensitisation and development of IgE-mediated food allergies.5
Several cohort studies have revealed that only 14–56% of patients harbour FLG gene null variants as a predisposing factor.6 , 7 The profilaggrin/FLG gene resides on chromosome 1q21.3 within the epidermal differentiation complex and consists of three exons. Exon 3 is extremely large (>12 kb) and sequencing is challenging as it encodes most of the profilaggrin polypeptide with almost complete homologous 10, 11 and 12 repeats.5 Null variants in exon 3 result in complete loss of the expressed protein. Every race is likely to have a unique set of FLG variants which could predict the severity of the disease.4 , 8 Most of the studies have analysed the more common loss of function variants, which could have resulted in under-reporting of other significant loss of function variants seen in different ethnic populations. Recent technology like next-generation sequencing can analyse the entire coding region of FLG to identify unreported loss of function variants.3
A recent Indian study has shown a prevalence of variant R501X to be 2.2% in children concomitantly associated with asthma and atopic dermatitis.9 Handa et al. have reported a prevalence of 33.7% of FLG variants (S2889X, 2282del4, R501X and Q2417X) associated with hand eczema from the Indian population.10 FLG variants could predict the outcome of atopic dermatitis which would help clinicians to counsel parents about the likely severity of atopic dermatitis. We aimed to study the FLG gene polymorphisms in Indian children with atopic dermatitis and do a genotype-phenotype correlation.
Materials and methods
Study population
This was a, cross-sectional, multicentre study of atopic dermatitis cases over 2 years, from April 2018 through April 2020. It was conducted at two centres in India, the Department of Pediatric Dermatology, Indira Gandhi Institute of Child Health, Bengaluru, Karnataka and the Institute of Child Health, Kolkata, West Bengal. We calculated the required sample size to be 69, based on the prevalence of FLG gene mutations in atopic dermatitis of 31.4 per cent.11 , 12 There has been no previous study in the Indian population available for comparison.
Children less than 18 years of age diagnosed with atopic dermatitis, with parents and all four grandparents of Indian ethnicity, presenting to the outpatient department of the two hospitals were enrolled. Children with mixed ethnicities were excluded. The diagnosis of atopic dermatitis was based on the criteria by Hanifin and Rajka.13 Demographic details were recorded on a predesigned proforma. Children were classified into three age groups based on the onset of atopic dermatitis: infantile (birth to 1 year), early childhood (1–8 years) and late childhood (8–18 years). Disease severity was assessed using the SCORing Atopic Dermatitis (SCORAD) index and classified as mild (<25), moderate (25–40) and severe (>40).14 Associated diseases like allergic rhinitis and asthma were diagnosed based on the questionnaire and previous diagnoses by a paediatrician. Written informed assent from the children and consent from the parents/guardians were obtained for clinical and molecular analysis. Institutional ethical committee clearances were taken from both the centres where the study was conducted (IEC/9/2018-19, IEC/183/2019).
Molecular analysis
Genomic DNA was extracted from peripheral blood (n = 75) using the genomic DNA extraction kit (Qiagen). DNA samples were amplified by polymerase chain reaction to different amplicon sizes to cover the coding regions (∼13.26 kb) of the FLG gene (three exons) using the specific primers. The amplicons were sheared (ME220, Covaris), pooled and libraries prepared (NEBNext kit-E7370L, NEB) as per the manufacturer’s protocol. The libraries were sequenced (HiSeq X, Illumina) to an average sequencing depth of ≥500x. A minimum of 90% of the sequenced bases were ≥Q30 value. The variants were called using Genome Analysis Toolkit (GATK) best practices pipeline annotated using a framework for the identification of variants in the sample using Sentieon (v201808.01). The sequences obtained were aligned to the human reference genome (GRCh37/hg19) using Sentieon aligner and analysed using Sentieon for removing duplicates, recalibration and realignment of indels. Gene annotation of the variants was performed using a variant effect predictor program against the Ensembl release 91 human gene model.15 Clinically relevant variants were annotated using published variants in literature and a set of diseases databases—ClinVar, OMIM, GWAS, HGMD (v2018.3) and SwissVar. Common variants were filtered based on allele frequency in 1000 Genome Phase 3, ExAC (v1.0), gnomAD (v2.1), EVS, dbSNP (v151), 1000 Japanese Genome and our internal Indian population database and in silico prediction tools [PolyPhen 2, Sorting Intolerant From Tolerant (SIFT), Mutation Taster2 and likelihood ratio test (LRT)]. The variants were analysed using Varminer (in-house variant interpretation tool) and prioritised. Around 100 age and sex matched controls were selected among the samples received for clinical exome sequencing for some other condition.
Statistical analysis
Descriptive statistics for quantitative values were expressed as means (± standard deviation) in accordance with the data distribution. Frequencies and percentages were used to describe categorical variables. Fisher’s exact test was used to compare parameters between atopic dermatitis with FLG variants and atopic dermatitis without FLG variants, as well as atopic dermatitis-associated phenotypes that included gender, family history of atopy, allergic disease association, pityriasis alba, palmar hyperlinearity, xerosis, cheilitis, SCORAD; and a Chi-square test for comparing the age of onset. P value was obtained from Student’s unpaired t-test for age at presentation. The P value for the significance of IgE was obtained from Mann–Whitney U test. The association between the occurrence of mutation and various phenotypes and clinical features was obtained by Spearman’s rank correlation (rho) and expressed along with confidence intervals. A similar Spearman’s rank correlation was done to interpret the association between the number of variants (single or more) and various phenotypic expressions and clinical parameters. MedCalc version 10.2 (MedCalc Software, 2011, Mariakerke, Belgium) was used for statistical analysis. A ‘P’ value of less than 0.05 (P < 0.05) was considered statistically significant.
Results
Clinical characteristics of study children
We recruited 80 unrelated children diagnosed with atopic dermatitis in the study. Five children failed the DNA quality control (insufficient DNA sample), hence we analysed 75 children [Figure 1]. Among these 75 children, 50 were boys and 25 were girls aged 4 months to 18 years (mean age, 5.2 ± 3.9 years). The clinical characteristics of children with and without FLG variants are presented in Table 1. The mean age of onset of the disease was 2.7 years (10 days to 12 years). Around 93.4% of children presented with atopic dermatitis in the early age group (birth to 8 years). Seventy children (93.3%) had moderate to severe disease in our study. In the present cohort, palmar hyperlinearity was seen in 42.7% and xerosis in 76% of children. Serum IgE levels could be done in only 55 children due to logistical reasons.
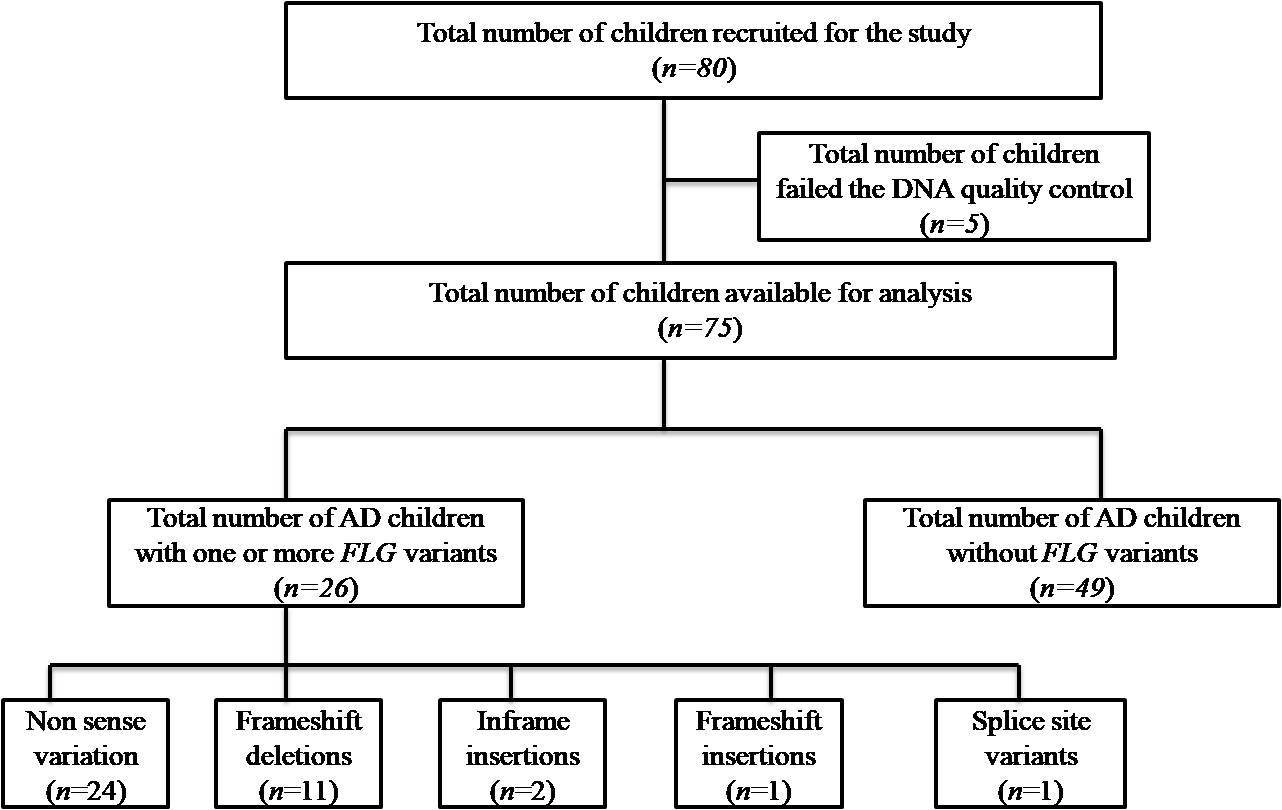
- Flow chart of the study
| Clinical features | AD with FLG variants (n = 26) | AD without FLG variants (n = 49) | Total (n = 75) | P |
|---|---|---|---|---|
| Age at enrolment (years) | ||||
| Mean ± SD | 4.57 ± 3.97 | 5.61 ± 3.88 | 5.25 ± 3.92 | 0.277 |
| Range | 0.4–13 | 0.4–18 | 0.4–18 | |
| Median (IQR) | 3.2 (1.8, 6) | 5 (2.45, 8.13) | 4 (2, 8) | |
| Gender | ||||
| Boys | 16 (61.5%) | 34 (69.4%) | 50 (66.7%) | 0.608 |
| Girls | 10 (38.4%) | 15 (30.6%) | 25 (33.3%) | |
| Age of onset | ||||
| Infantile (birth–<1 year) | 17 (65.4%) | 18 (36.7%) | 35 (46.7%) | 0.041 |
| Early childhood (1–8 years) | 7 (26.9%) | 28 (57.1%) | 35 (46.7%) | |
| Late childhood (8–18 years) | 2 (7.7%) | 3 (6.1%) | 5 (6.7%) | |
| Family history | 10 (38.5%) | 22 (44.9%) | 32 (42.7%) | 0.632 |
| Allergic disease association (allergic rhinitis and/or asthma) | 10 (38.5%) | 24 (49.0%) | 34 (45.3%) | 0.468 |
| SCORAD | ||||
| Mild (<15) | 1 (3.8%) | 4 (8.2%) | 5 (6.7%) | 0.653 |
| Moderate (15–40) | 16 (61.5%) | 35 (71.4%) | 51 (68%) | 0.440 |
| Severe (>40) | 9 (34.6%) | 10 (24.4%) | 19 (25.3%) | 0.264 |
| Pityriasis alba | 6 (23.1%) | 14 (28.6%) | 20 (26.7%) | 0.785 |
| Palmar hyperlinearity | 12 (46.2%) | 20 (40.8%) | 32 (42.7%) | 0.807 |
| Xerosis | 21 (80.8%) | 36 (73.5%) | 57 (76%) | 0.577 |
| Cheilitis | 3 (11.5%) | 12 (24.5%) | 15 (20%) | 0.234 |
P value <0.05 was considered significant. SD, Standard deviation; IQR, Interquartile range; AD, Atopic dermatitis; FLG, Filaggrin gene; SCORAD: SCORing Atopic Dermatitis
Molecular results
This study documented 26 children having one or more FLG null variants. The prevalence of FLG loss of function mutation was 34.7%. The data on FLG loss of function variants in our study are summarised in Table 2. There were 20 different FLG loss of function variants in our study. Of these 24 (61%) were nonsense mutations, 11(28%) frameshift deletions, 1(3%) frameshift insertions, 1(3%) splice loss of function variants and 2(5%) inframe insertions. Among the 20 FLG loss of function, 16 (80%) were unique to our study, not reported in any database or literature [Table 3]. All these novel variants were not detected in 100 people who did not have FLG variants. Four variants (20%) in ten patients in our study were previously reported null variants (shown in bold in Table 2: p.Arg2447Ter, p.Arg501Ter, c.2282del4 and p.Arg2037Ter). Seven variants were detected in more than one individual of whom four were novel (p. Ser3640Ter, p.Ser2344Ter, p.Arg992SerfsTer31 and p.Trp1064Ter) and three (p.Ser761CysfsTer36, p.Arg2447Ter and p.Arg501Ter) were previously reported null variants. Two variants, nonsense (p. Ser3640Ter) and frameshift (p.Ser761CysfsTer36) were detected in five patients each and another nonsense variant (p.Ser2344Ter) in four patients. Three (11.5%) children had homozygous null variants (p.Ser761cysfsTer36, p.Arg99SerfsTer31 and p.Arg501Ter) and 23 (88.5%) children had heterozygous null variants. Among the 13 children with more than one variant, 11 (42.3%) had two null variants and two (7.7%) had three null variants.
Previously reported null variants are highlighted in bold. VarClass, variant classifier; cDNA, complementary DNA; AA, amino acid; dbSNP ID, single nucleotide polymorphism database; FRAMESHIFT-DEL, frameshift deletion; FRAMESHIFT-INS, frameshift insertion VarClass, variant classifier; cDNA, complementary DNA; AA, amino acid; dbSNP ID, single nucleotide polymorphism database; FRAMESHIFT-DEL, frameshift deletion; FRAMESHIFT-INS, frameshift insertion; gnomAD, genome aggregation database
Patient ID
Transcript ID
VarClass
cDNA change
AA change
Zygosity
dbSNP ID
Variant
237870
NM_002016.2
NONSENSE
c.1339C>T
p.Gln447Ter
Heterozygous
rs763028697
chr1:152286023G>A
237870
NM_002016.2
INTRONIC-SS-ACR
c.139-1G>A
NA
Heterozygous
−
chr1:152287224C>T
288486
NM_002016.2
NONSENSE
c.6109C>T
p.Arg2037Ter
Heterozygous
rs200002200
chr1:152281253G>A
303109
NM_002016.2
NONSENSE
c.7339C>T
p.Arg2447Ter
Heterozygous
rs138726443
chr1:152280023G>A
196655
NM_002016.2
NONSENSE
c.7339C>T
p.Arg2447Ter
Heterozygous
rs138726443
chr1:152280023G>A
327946
NM_002016.2
NONSENSE
c.1501C>T
p.Arg501Ter
Heterozygous
rs61816761
chr1:152285861G>A
261722
NM_002016.2
NONSENSE
c.1501C>T
p.Arg501Ter
Homozygous
rs61816761
chr1:152285861G>A
237872
NM_002016.2
NONSENSE
c.1501C>T
p.Arg501Ter
Heterozygous
rs61816761
chr1:152285861G>A
303026
NM_002016.2
NONSENSE
c.1714C>T
p.Arg572Ter
Heterozygous
rs200601767
chr1:152285648G>A
303026
NM_002016.2
FRAMESHIFT-DEL
c.2976_2977del
p.Arg992SerfsTer31
Heterozygous
rs776968118
chr1:152284384GCT>G
261725
NM_002016.2
FRAMESHIFT-DEL
c.2976_2977del
p.Arg992SerfsTer31
Heterozygous
rs776968118
chr1:152284384GCT>G
254989
NM_002016.2
FRAMESHIFT-DEL
c.2976_2977del
p.Arg992SerfsTer31
Homozygous
rs776968118
chr1:152284384GCT>G
327941
NM_002016.2
FRAMESHIFT-DEL
c.5014del
p.Gln1672ArgfsTer34
Heterozygous
rs771090956
chr1:152282347TG>T
261717
NM_002016.2
NONSENSE
c.6940C>T
p.Gln2314Ter
Heterozygous
rs762689918
chr1:152280422G>A
237856
NM_002016.2
NONSENSE
c.9315C>G
p.Tyr3105Ter
Heterozygous
rs200815866
chr1:152278047G>C
227344
NM_002016.2
FRAMESHIFT-INS
c.4808_4812dup
p.Glu1605ThrfsTer103
Heterozygous
rs775716153
chr1:152282549C>CTGAGT
303034
NM_002016.2
FRAMESHIFT-DEL
c.6067_6082del
p.Gly2023HisfsTer67
Heterozygous
−
chr1:152281279GAAGCTTGTCCATGCCC>G
227335
NM_002016.2
NONSENSE
c.7777G>T
p.Gly2593Ter
Heterozygous
rs756353784
chr1:152279585C>A
196661
NM_002016.2
NONSENSE
c.1279G>T
p.Gly427Ter
Heterozygous
rs544327834
chr1:152286083C>A
237894
NM_002016.2
FRAMESHIFT-DEL
c.8590_8591del
p.His2864CysfsTer5
Heterozygous
rs1407703398
chr1:152278770ATG>A
343834
NM_002016.2
NONSENSE
c.7031C>G
p.Ser2344Ter
Heterozygous
rs372754256
chr1:152280331G>C
343833
NM_002016.2
NONSENSE
c.7031C>G
p.Ser2344Ter
Heterozygous
rs372754256
chr1:152280331G>C
256713
NM_002016.2
NONSENSE
c.7031C>G
p.Ser2344Ter
Heterozygous
rs372754256
chr1:152280331G>C
254986
NM_002016.2
NONSENSE
c.7031C>G
p.Ser2344Ter
Heterozygous
rs372754256
chr1:152280331G>C
343834
NM_002016.2
NONSENSE
c.10919C>G
p.Ser3640Ter
Heterozygous
rs745827275
chr1:152276443G>C
343833
NM_002016.2
NONSENSE
c.10919C>G
p.Ser3640Ter
Heterozygous
rs745827275
chr1:152276443G>C
327948
NM_002016.2
NONSENSE
c.10919C>G
p.Ser3640Ter
Heterozygous
rs745827275
chr1:152276443G>C
256713
NM_002016.2
NONSENSE
c.10919C>G
p.Ser3640Ter
Heterozygous
rs745827275
chr1:152276443G>C
254986
NM_002016.2
NONSENSE
c.10919C>G
p.Ser3640Ter
Heterozygous
rs745827275
chr1:152276443G>C
343830
NM_002016.2
FRAMESHIFT-DEL
c.2282_2285del
p.Ser761CysfsTer36
Heterozygous
rs558269137
chr1:152285076CACTG>C
288486
NM_002016.2
FRAMESHIFT-DEL
c.2282_2285del
p.Ser761CysfsTer36
Heterozygous
rs558269137
chr1:152285076CACTG>C
261725
NM_002016.2
FRAMESHIFT-DEL
c.2282_2285del
p.Ser761CysfsTer36
Heterozygous
rs558269137
chr1:152285076CACTG>C
254979
NM_002016.2
FRAMESHIFT-DEL
c.2282_2285del
p.Ser761CysfsTer36
Homozygous
rs558269137
chr1:152285076CACTG>C
227339
NM_002016.2
FRAMESHIFT-DEL
c.2282_2285del
p.Ser761CysfsTer36
Heterozygous
rs558269137
chr1:152285076CACTG>C
343834
NM_002016.2
NONSENSE
c.3191G>A
p.Trp1064Ter
Heterozygous
rs546421276
chr1:152284171C>T
343833
NM_002016.2
NONSENSE
c.3191G>A
p.Trp1064Ter
Heterozygous
rs546421276
chr1:152284171C>T
227344
NM_002016.2
NONSENSE
c.7434C>G
p.Tyr2478Ter
Heterozygous
rs144157090
chr1:152279928G>C
Patient ID
VarClass
cDNA change
AA change
Allele frequency
gnomAD
dbSNP ID
237870
INTRONIC-SS-ACR
c.139-1G>A
NA
0.006666667
NA
−
303026
NONSENSE
c.1714C>T
p.Arg572Ter
0.006666667
0.00009746
rs200601767
327941
FRAMESHIFT-DEL
c.5014del
p.Gln1672ArgfsTer34
0.006666667
0.00002843
rs771090956
261717
NONSENSE
c.6940C>T
p.Gln2314Ter
0.006666667
0.00001218
rs762689918
237856
NONSENSE
c.9315C>G
p.Tyr3105Ter
0.006666667
0.00001361
rs200815866
237870
NONSENSE
c.1339C>T
p.Gln447Ter
0.006666667
0.00000406
rs763028697
303034
FRAMESHIFT-DEL
c.6067_6082del
p.Gly2023HisfsTer67
0.006666667
NA
−
227335
NONSENSE
c.7777G>T
p.Gly2593Ter
0.006666667
0.00000812
rs756353784
196661
NONSENSE
c.1279G>T
p.Gly427Ter
0.006666667
0.00000812
rs544327834
343834, 343833, 327948, 256713, 254986
NONSENSE
c.10919C>G
p.Ser3640Ter
0.033333333
0.00006558
rs745827275
343834, 343833
NONSENSE
c.3191G>A
p.Trp1064Ter
0.013333333
0.00015025
rs546421276
227344
NONSENSE
c.7434C>G
p.Tyr2478Ter
0.006666667
0.00001218
rs144157090
303026, 261725, 254989
FRAMESHIFT-DEL
c.2976_2977del
p.Arg992SerfsTer31
0.026666667
0.0001868
rs776968118
227344
FRAMESHIFT-INS
c.4808_4812dup
p.Glu1605ThrfsTer103
0.006666667
0.00000812
rs775716153
237894
FRAMESHIFT-DEL
c.8590_8591del
p.His2864CysfsTer5
0.006666667
NA
rs1407703398
343834, 343833, 256713, 254986
NONSENSE
c.7031C>G
p.Ser2344Ter
0.026666667
0.0004731
rs372754256
Association of FLG null variants with atopic dermatitis
In our study FLG variants were significantly associated with early age of onset (P = 0.016) and elevated serum IgE levels which just reached statistical significance (P = 0.051) [Table 4]. About 65.4% of children with FLG variants presented with atopic dermatitis in the infantile age group. Though 96% of atopic dermatitis children with FLG variants had moderate to severe atopic dermatitis, it was not a statistically significant finding (P = 0.154). A negative correlation between pityriasis alba and FLG variants was found [Table 4]. Clinical characteristics of 13 (50%) children who had two or more FLG null variants are described in Table 5. No significant associations were observed between the FLG genotype with two or more null variants and severity or other atopic dermatitis-associated phenotypes, except the early age of onset [Table 6].
NA, Not applicable
Clinical features
Correlation coefficient (rho)
Confidence interval
P
Age of onset of atopic dermatitis (months)
−0.277
−0.475 to –0.0539
0.016
Family history of atopy
0.0619
−0.167 to 0.285
0.598
SCORAD severity
0.166
−0.063 to 0.379
0.154
Allergic disease association (allergic rhinitis and/or asthma)
−0.0997
−0.319 to 0.130
0.395
Serum IgE levels (IU/mL)
0.264
−0.001 to 0.495
0.051
Pityriasis alba
−0.276
−0.473 to −0.0524
0.0165
Palmar hyperlinearity
0.0514
−0.178 to 0.275
0.6617
Xerosis
0.0813
−0.148 to 0.303
0.4878
Cheilitis
−0.154
−0.368 to 0.0755
0.1869
Patient ID
Variant
Variant_1
Variant_2
Variant_3
Age of onset
Family history
Allergic disease association
SCORAD
Other features
227344
Two heterozygous variants
p.Glu1605ThrfsTer103
p.Tyr2478Ter
NA
10 days
Wheezing
Nil
Moderate
Xerosis
237856
Two heterozygous variants
p.Gln2659_Gln2660insLeu
p.Tyr3105Ter
NA
1 month
Nil
Nil
Moderate
Xerosis/hyperlinearity
237870
Two heterozygous variants
c.139-1G>A
p.Gln447Ter
NA
3 months
Nil
Nil
Severe
Xerosis/hyperlinearity/cheilitis
254979
Homozygous
p.Ser761CysfsTer36
p.Ser761CysfsTer36
NA
6 months
Nil
Nil
Moderate
Xerosis/hyperlinearity
254986
Two heterozygous variants
p.Ser2344Ter
p.Ser3640Ter
NA
6 months
Nil
Nil
Moderate
Xerosis/hyperlinearity
254989
Homozygous
p.Arg992SerfsTer31
p.Arg992SerfsTer31
NA
6 months
Nil
Allergic rhinitis
Moderate
Xerosis/hyperlinearity/periorbital darkening
256713
Two heterozygous variants
p.Ser2344Ter
p.Ser3640Ter
NA
5 months
Atopic dermatitis
Nil
Moderate
Xerosis/hyperlinearity
261722
Homozygous
p.Arg501Ter
p.Arg501Ter
NA
12 years
asthma
Allergic rhinitis
Moderate
Nil
261725
Two heterozygous variants
p.Arg992SerfsTer31
p.Ser761CysfsTer36
NA
3 months
Nil
Allergic rhinitis
Severe
Xerosis
288486
Two heterozygous variants
p.Arg2037Ter
p.Ser761CysfsTer36
NA
4 months
Wheezing
Nil
Moderate
Xerosis/hyperlinearity
303026
Two heterozygous variants
p.Arg572Ter
p.Arg992SerfsTer31
NA
10 days
Asthma
Allergic rhinitis
Severe
Xerosis
343833
Three heterozygous variants
p.Ser2344Ter
p.Ser3640Ter
p.Trp1064Ter
3 months
Nil
Nil
Moderate
Keratosis pilaris
343834
Three heterozygous variants
p.Ser2344Ter
p.Ser3640Ter
p.Trp1064Ter
3 months
Nil
Nil
Mild
Nil
Clinical features
Correlation coefficient (rho)
Confidence interval
P
Age of onset of atopic dermatitis (months)
−0.614
−0.809 to −0.297
0.0008
Family history of atopy
0.00
–
1.00
SCORAD severity
−0.277
−0.600 to 0.124
0.1711
Allergic disease association (allergic rhinitis and/or asthma)
−0.203
−0.547 to 0.200
0.3203
Serum IgE levels (IU/mL)
−0.208
−0.596 to 0.258
0.3782
Pityriasis alba
−0.548
−0.771 to −0.204
0.0038
Palmar hyperlinearity
0.154
−0.248 to 0.511
0.4517
Xerosis
−0.0976
−0.467 to 0.301
0.6353
Cheilitis
−0.120
−0.485 to 0.280
0.5580
Discussion
There are more than 500 stop-gain variants in FLG described in the genome aggregation database.16 The most common FLG variants shared among Japanese, Koreans, Taiwanese, Chinese, Singaporeans and Asians are R501X and c.3321delA. In the European population, the most prevalent mutations are R501X and c.2282del4. The three most common FLG variants R501X, c.2282del4 and E2422X are shared in both European and Asian populations.8 Meng et al. found c.3321delA to be associated with xerosis, ichthyosis vulgaris, palmar hyperlinearity, keratosis pilaris, white dermographism and disease severity but not associated with early age of onset.17 Previous studies found that FLG loss of function variants were not commonly seen in persons of African ancestry, which could be due to inappropriate genotyping techniques. Recent studies with massively parallel sequencing have shown uncommon FLG variants to be associated with persistent atopic dermatitis in African ancestry as compared to European ancestry. However, the frequency of FLG loss of function mutations in African ancestry is lower. Copy number variation has been associated as a risk factor for atopic dermatitis and this has been documented in African-American pediatric patients with moderate to severe atopic dermatitis.18
About 20 FLG loss of function variants in 26 children (6 frameshift, 13 nonsense and 1 splice site variant) were seen in our study.
Among the documented FLG loss of function variants in our study, 20% of the variants were previously reported in European and Asian populations and 80% were unique variants.16 , 17 , 19 , 20 India is home to more than 2000 ethnic populations, whose genetic origins are complex. The present Indian population is genetically heterogeneous with an admixture of ancestral north Indians comprising the majority and having roots in the Middle East, Central Asia and Europe and ancestral south Indians that have no relation to any population outside India.21 Further complexity is added by the relatively recent immigration of populations such as Siddis, Muslims and Jews.22 Ours being a two-centre study, had both these sets of populations (East Indians and South Indians) and thus reported variants of Asia and European as well as a high number of unique variants. Chauhan et al. studied the prevalence of R501X mutation in Indian children with allergic diseases. Their study was a case-control study study enrolling 90 children with allergic diseases (asthma, rhinitis and atopic dermatitis). The mutant R501X was seen in 3.3% of children with asthma and 2.2% in children with asthma concomitantly with eczema. The limitation of their study was that screening of only one FLG null variant was done.9
Several studies have shown the strong association of FLG null mutation with different atopic dermatitis phenotypes.16 FLG null variants in our study were significantly associated with early age of onset (infantile) and elevated serum IgE levels, which was concordant with several earlier studies.23 , 24 , 25 FLG variants have a significant effect on the age of onset of atopic dermatitis and can lead to early onset (less than 1 year) and persistence of the disease into adulthood.23 , 24 Although previous studies have shown a significant association between the FLG null variants with moderate to severe atopic dermatitis (odds ratio 2.03–13.4), our study did not show a statistically significant correlation.4 , 11 , 26 , 27 FLG mutation is a strong genetic predisposing factor for atopic dermatitis, but other factors like unidentified candidate genes, copy number variations and epigenetic factors may also play an important role in the pathogenesis of atopic dermatitis, which could explain the non-correlation of the severity of atopic dermatitis with FLG variants in our study, as well as the fact that SCORAD is variable at different points of time.16 Interestingly Park et al. in a study on Korean patients with atopic dermatitis did not observe any significant association between FLG null variants and clinical features of atopic dermatitis.18
Ota et al. studied the effect of FLG loss of function mutation on the severity of skin lesions and skin barrier function in the Japanese population. Eight (14.5%) patients had a loss of function mutations in the FLG gene in their study. Stratum corneum was collected from three different sites (extremities, neck and trunk) using the tape-stripping method. The amount of FLG protein and total amino acid in the stratum corneum was measured in both mutation carriers and non-carriers. FLG abnormalities had little effect on the severity of dermatitis, FLG protein and total amino acid content in the stratum corneum in the lesional skin. Their study suggested that the activation of Th2 dominant inflammatory cells along with FLG abnormalities plays a role in suppressing the production of FLG in lesional skin.28
Previous studies have shown FLG mutation to be associated with elevated levels of IgE in Japanese, Koreans, Singaporeans, Chinese and European populations.4 , 29 In our study too, serum IgE levels were significantly associated with FLG null variants. We were unable to find any association between FLG variants with allergic disease association, family history, palmar hyperlinearity, xerosis, cheilitis and keratosis pilaris and this was similar to other studies.4 , 7 , 26
We found no significant associations in severity or clinical manifestations of atopic dermatitis between children carrying one single variant and children with two or more variants. This could again be explained by the multifactorial nature of the disease. Though FLG mutation is not an independent risk factor for the development of different atopic dermatitis phenotypes, studies on FLG variants may help identify some infants at risk of developing atopic dermatitis, guiding early interventions to delay the onset and decrease the severity of atopic dermatitis.
Limitations
The main limitation of our study is its relatively small sample size. As a result, there could have been false negative or positive results due to low statistical power. The small sample size in our study also did not permit a statistical correlation between specific null variants and atopic dermatitis phenotypes. Larger population-based studies are required to substantiate the associations between FLG mutations and atopic dermatitis phenotypes in the Indian population. We could not look for copy number variation in the present study due to logistic reasons.
Conclusion
Our study documented a high prevalence of FLG null variants in Indian children. The FLG variants identified in our study included ones unique to this population and also those that overlapped with Asian and European populations. The presence of more than one FLG mutations in an individual did not correlate significantly with the severity of AD. We found a significant association of FLG variants with an early age of onset and elevated serum IgE levels.
Acknowledgements
The authors would like to thank the Indian Society for Paediatric Dermatology for funding our research project and all the patients for participating in the study.
Declaration of patient consent
The authors certify that they have obtained all appropriate patient consent.
Financial support and sponsorship
Indian Society of Paediatric Dermatology
Conflicts of interest
There are no conflicts of interest
References
- Atopic dermatitis: Global epidemiology and risk factors. Ann NutrMetab. 2015;66:8-16.
- [CrossRef] [PubMed] [Google Scholar]
- Atopic dermatitis in infants and children in India. Indian J Dermatol VenereolLeprol. 2010;76:504-13.
- [CrossRef] [PubMed] [Google Scholar]
- The role of filaggrin in atopic dermatitis and allergic disease. Ann Allergy Asthma Immunol. 2020;124:36-43.
- [CrossRef] [PubMed] [Google Scholar]
- Filaggrin mutation in Korean patients with atopic dermatitis. Yonsei Med J. 2017;58:395-400.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Filaggrin gene defects and the risk of developing allergic disorders. Allergol Int. 2011;60:1-9.
- [CrossRef] [PubMed] [Google Scholar]
- Filaggrin gene defects and risk of developing allergic sensitisation and allergic disorders: Systematic review and meta-analysis. BMJ. 2009;339:B2433.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Wide spectrum of filaggrin-null mutations in atopic dermatitis highlights differences between Singaporean Chinese and European populations. Br J Dermatol. 2011;165:106-14.
- [CrossRef] [PubMed] [Google Scholar]
- Filaggrin gene polymorphisms in Iranian ichthyosis vulgaris and atopic dermatitis patients. Int J Dermatol. 2018;57:1485-91.
- [CrossRef] [PubMed] [Google Scholar]
- Prevalence of filaggrin gene R501X mutation in Indian children with allergic diseases. Indian J Pediatr. 2020;87:587-90.
- [CrossRef] [PubMed] [Google Scholar]
- Filaggrin gene mutations in hand eczema patients in the Indian subcontinent: A prospective case-control study. Contact Dermatitis. 2019;80:359-64.
- [CrossRef] [PubMed] [Google Scholar]
- Mutations in the filaggrin gene in Han Chinese patients with atopic dermatitis. Allergy. 2011;66:420-7.
- [CrossRef] [PubMed] [Google Scholar]
- Sample size calculation in medical studies. Gastroenterol Hepatol Bed Bench. 2013;6:14-17.
- [PubMed] [PubMed Central] [Google Scholar]
- Consensus Report of the European Task Force on Atopic Dermatitis. Dermatology. 1993;186:23-31.
- [CrossRef] [PubMed] [Google Scholar]
- Ensembl 2018. Nucleic Acids Res. 2018;46:D754-61.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Filaggrin gene mutations with special reference to atopic dermatitis. Curr Treat Options Allergy. 2020;7:403-13.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Filaggrin gene mutation c.3321delA is associated with various clinical features of atopic dermatitis in the Chinese Han population. PLoS One. 2014;9:e98235.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Novel FLG null mutations in Korean patients with atopic dermatitis and comparison of the mutational spectra in Asian populations. J Dermatol. 2015;42:867-73.
- [CrossRef] [PubMed] [Google Scholar]
- X-linked and autosomal dominant forms of the ichthyosis in coinheritance. Drug Metab Pers Ther 2019:34.
- [CrossRef] [PubMed] [Google Scholar]
- Genetic evidence for recent population mixture in India. Am J Hum Genet. 2013;93:422-38.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Complex genetic origin of Indian populations and its implications. J Biosci. 2012;37:911-9.
- [CrossRef] [PubMed] [Google Scholar]
- Uncommon Filaggrin Variants Are Associated with Persistent Atopic Dermatitis in African Americans. J Invest Dermatol. 2018;138:1501-1506.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Clinical characteristics of Korean patients with filaggrin-related atopic dermatitis. Clin Exp Dermatol. 2016;41:595-600.
- [CrossRef] [PubMed] [Google Scholar]
- Null mutations in the filaggrin gene (FLG) determine major susceptibility to early-onset atopic dermatitis that persists into adulthood. J Invest Dermatol. 2007;127:564-7.
- [CrossRef] [PubMed] [Google Scholar]
- The atopic march: Progression from atopic dermatitis to allergic rhinitis and asthma. J Clin Cell Immunol. 2014;5:202.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Filaggrin loss-of-function mutations as a predictor for atopic eczema, allergic sensitization and eczema-associated asthma in Polish children population. Adv Clin Exp Med. 2017;26:991-8.
- [CrossRef] [PubMed] [Google Scholar]
- Filaggrin null mutations and childhood atopic eczema: A population-based case-control study. J Allergy Clin Immunol. 2008;121:940-46.e3.
- [CrossRef] [PubMed Central] [Google Scholar]
- Filaggrin-gene mutation has minimal effect on the disease severity in the lesions of atopic dermatitis. J Dermatol. 2021;48:1688-1699.
- [CrossRef] [PubMed] [Google Scholar]
- Filaggrin null mutations are associated with atopic dermatitis and elevated levels of IgE in the Japanese population: A family and case-control study. J Hum Genet. 2008;53:615-621.
- [CrossRef] [PubMed] [Google Scholar]






