Translate this page into:
Investigating the link between toll-like receptor 9 gene variants and vitiligo susceptibility - A cross-sectional comparative study
Corresponding author: Dr. Rasha Turky Abdel-Razek Abdel-Aziz, Department of Dermatology, Sexually Transmitted Diseases and Andrology, Faculty of Medicine, Minia University, Minia, Egypt. Email address: rashatorky@yahoo.com.
-
Received: ,
Accepted: ,
How to cite this article: Abdel-Aziz RTA, Hammad SS, Ahmed SS. Investigating the link between toll-like receptor 9 gene variants and vitiligo susceptibility - A cross-sectional comparative study. Indian J Dermatol Venereol Leprol. doi: 10.25259/IJDVL_1041_2024
Abstract
Background
The Toll-like receptor (TLR) family, which recognises diverse molecular patterns on immune cells, has been implicated in several autoimmune diseases, including vitiligo.
Objectives
This study will investigate the potential association between the TLR9 gene polymorphism rs187084 and clinical features in Egyptian vitiligo patients, employing the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method for TLR9 genotyping.
Methods
Genomic DNA was extracted from the peripheral blood of 30 vitiligo patients and 20 healthy controls using a DNA isolation kit. TLR9 rs187084 gene polymorphisms were assessed using PCR-RFLP techniques. Genotype distributions and allele frequencies were compared between vitiligo patients and controls. Additionally, the associations between TLR9 single nucleotide polymorphism (SNP) genotypes and clinical features including demographic characteristics, family history, disease onset, activity, duration, and hair involvement in vitiligo patients were statistically analysed.
Results
The CC genotype of rs187084, which is considered protective, was found to be more prevalent in the control group compared to the vitiligo group. However, no significant associations were identified between TLR9 SNP genotypes and demographic or clinical parameters, including family history, disease onset, activity, duration, and hair involvement (leukotrichia) in vitiligo patients.
Limitation
The limitations of this study include a small sample size and the methodological constraints of PCR-RFLP, necessitating further research to confirm our findings and elucidate underlying mechanisms.
Conclusions
The significant difference in the distribution of the TLR9 rs187084 polymorphism between vitiligo patients and controls highlights the potential role of innate immunity in the susceptibility to vitiligo. Despite the absence of significant associations with clinical parameters in this study, these findings underscore the importance of genetic factors in the pathogenesis of vitiligo.
Keywords
Vitiligo
autoimmunity
innate immunity
TLR9
RFLP
Introduction
Vitiligo is an acquired, chronic disorder characterised by depigmented patches with an unknown aetiology and a polygenic inheritance pattern.1 It is a complex disease influenced by genetic and environmental factors.2 Some researchers suggest it is a multifactorial disorder with numerous genetic causes.3
Various pathophysiological mechanisms have been proposed for vitiligo, including autoimmune, neural, autocytotoxic, biochemical, oxidative stress, melanocytorrhagy, and decreased melanocyte survival.4
The autoimmune theory is particularly relevant in generalized vitiligo, which is considered a complex disorder involving the combined pathogenic effects of multiple susceptibility genes and unidentified environmental factors, resulting in the autoimmune destruction of melanocytes.4 Vitiligo is a polygenic disorder, with many genes related to autoimmunity implicated in its pathogenesis, each genetic factor making a small individual contribution.5,6 However, the polygenicity of vitiligo is low, and its heritability is high when compared to most other complex traits, which might make its genetic architecture easier to discover.2
Vitiligo serves as a model for investigating the genetic architecture of complex diseases and could offer significant insights for predictive and personalized medicine in complex diseases.7 The role of innate immunity in vitiligo is evident by the infiltration of natural killer (NK) cells, which are five times more prevalent in the lesional skin of vitiligo patients when compared to healthy controls, and almost two-fold in unaffected skin.8 TLRs, identified in humans in 1998, are components of innate immunity that recognise pathogen-associated molecular patterns (PAMPs) present in microbes.9 TLRs function by binding to lipoproteins and peptidoglycans (TLR2, 4, 5, 6, and 11), or nucleic acid ligands associated with viral and bacterial single-strand RNA or double-strand RNA (TLR3, 7, 8, and 9). Upon ligand binding, TLRs participate in pathogen eradication through inflammatory cytokines and co-stimulatory gene expression.10
TLRs are the most well-characterised pattern recognition receptors (PRRs) and are transmembrane proteins encoded by the toll gene family.4 They are expressed in various cell types, including immune and nonimmune cells, and play a crucial role in detecting PAMPs and damage-associated molecular patterns (DAMPs). They also activate the adaptive immune system through the upregulation of costimulatory molecules on antigen-presenting cells (APCs).9
The signalling mediated by endogenous TLR ligands is crucial in autoimmune disorders.11 Activation of certain TLRs through DAMP interactions can facilitate tissue repair and the elimination of cell debris; however, this same interaction is linked to the chronic inflammatory processes involved in rheumatologic diseases.12 Therefore, TLR activation by DAMPs appears to play a role in the self-sustained inflammatory cycles and progression of these chronic diseases.13
Single-nucleotide polymorphism (SNP) genotyping technologies investigate the effects of amino acid variations on protein function. SNPs in promoter regions can alter transcription factor binding motifs, suppress wild-type transcript production, modify enhancer or repressor element efficiency, or introduce alternative translation initiation codons.14
There is a paucity of research on TLR gene polymorphisms in Egyptian vitiligo patients. This study aims to determine whether there is an association between TLR9 polymorphisms, specifically rs187084, in Egyptian vitiligo patients and their clinical characteristics including family history, disease onset, activity, duration, and hair involvement.
Methods
The sample size was determined to provide 80% power for chi-square (X2) goodness-of-fit tests. The type of power analysis used was an a priori computation to determine the required sample size, based on a significance level of 0.05 and an effect size of 0.5. This calculation was performed using G*Power 3.1.9.7 software. The present study entailed a cross-sectional comparison comprising 50 participants in two groups:
Group 1: Consisted of 30 individuals clinically diagnosed with vitiligo, ascertained both through clinical evaluation by dermatologist and Wood’s light examination. These patients were recruited from our outpatient Department of Dermatology, STDs, and Andrology. Detailed medical histories encompassing personal information, disease progression, psychological trauma, other concurrent skin or systemic disorders, and familial predispositions to vitiligo or other autoimmune conditions were documented for each patient.
Group 2: Included twenty healthy subjects selected as a control cohort.
Ethical approval for the study was obtained from the local scientific research ethics committee at the Faculty of Medicine (Approval No. 652:2023). Written consent was taken from all patients and control prior to the study.
Skin assessments in Group 1 involved clinical inspection for vitiligo manifestations, with attention to lesion characteristics, distribution patterns, and associated comorbidities, validated by Wood’s light examination.
Vitiligo activity (appearance of new depigmented lesions or the enlargement of existing ones) was quantified using the Vitiligo Disease Activity Score (VIDA), employing a six-point scale reflective of disease progression over time. VIDA score is based on patients’ opinions and divided into six grades as follows 15
+4: Activity of 6 weeks or less preceding the assessment
+3: Activity of 6 weeks to 3 months
+2: Activity of 3 to 6 months
+1: Activity of 6 to 12 months
0: Stable for at least 1 year and
−1: Stable for at least 1 year with spontaneous regimentation
Genotyping
Blood specimens were collected in ethylene diamine tetra acetic acid (EDTA) tubes and stored at -20°C until processing. Genomic DNA was isolated from whole blood using the QIAamp DNA Mini Kit (cat no.51104, QIAGEN, Germany) following standard protocols.
Genotyping of SNPs at the TLR9 rs187084 locus was performed via polymerase chain reaction and restriction fragment length polymorphism (PCR-RFLP assay. Oligonucleotide primers specific to rs187084 were utilised:
F: 5’-TAT CGT CTT ATT CCC CTG CTGGAATGT -3’ (sense) and
R: 5-TGCCCAGAGCTGACTGCTGG-3’ (anti- sense).13
PCR conditions entailed initial denaturation at 95°C, followed by 35 cycles of denaturation for 30 seconds, annealing at 60°C for 25 seconds, and extension at 72°C for 40 seconds using a thermocycler (UNOII). Amplicons were then subjected to Afl II restriction endonuclease digestion at 37°C for 1 hour (Enzynomics, Cat. R042S, Korea), and the resulting fragments were visualised on an agarose gel stained with ethidium bromide. A DNA ladder with 50 bp (Sizer tm DNA Markers from QIAGEN, USA) was used as a marker.
Statistical analysis
Statistical analyses were conducted using IBM SPSS version 25 (IBM Corp. Released 2017. IBM SPSS Statistics for Windows, Version 25.0. Armonk, NY: IBM Corp). Data normality was assessed via the Kolmogov-Smirnov test and Brown-Frosythe test for median (0.232 for duration of disease and 0.342 for VIDA score), and it was noted that the duration of disease and VIDA score had homogenetic variance. Hence, the Kruskal Wallis test was used for these non-parametric quantitative data between three gene groups. The Chi-square test was used to compare cases and controls regarding genotype and phenotype, while the Fisher Exact test was used to compare family history, VIDA score, hair involvement, type of vitiligo and onset of disease between different genotype groups. Multivariable Logistic Regression analysis was used to analyse the effect of phenotype characteristics including gender, family history, age of onset, disease duration, type of vitiligo and disease activity on the dependent variable (allele frequency T and C) in cases. P-value less than 0.05 was considered statistically significant.
Results
Study design and population demographics
This was a cross-sectional comparative study involving 50 subjects sub-divided into two groups:
-
Group 1 comprised 30 vitiligo patients, 19 females (63.3%) and 11 males (36.7%). Their ages ranged from 8 to 80 years (mean ± SD, 35.9 ± 23.1 years). A positive family history was reported in 5 (16.7 %) patients. Disease activity using the VIDA score was distributed as in Figure 1, with a mean ± SD of 0.97 ± 1.6. The duration of the disease ranged from 1 to 37 years (mean ± SD, 6.9 ± 8.4 years). The patients included in the study had non-segmental vitiligo in 22 (73.3%) cases and segmental vitiligo in 8 (26.7%) cases. Early onset disease (before 12 years of age) was reported in 16 (53.3%) patients, while 14 cases (46.7%) showed late onset disease. Hair involvement was reported in only 5 (16.7%) cases, while the remaining 25 (83.3%) patients showed no hair involvement.
-
Group 2 consisted of 20 healthy controls, with 7 males (35%) and 13 females (65%). Their ages ranged from 8-70 years.
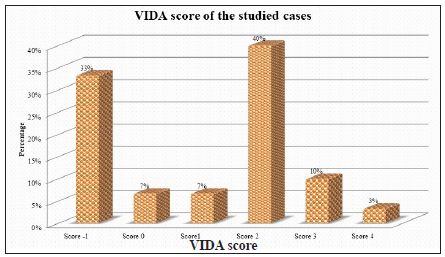
- Distribution of disease activity in vitiligo patients. (VIDA score: Vitiligo disease activity score)
Distribution of TLR9 rs187084 genotypes
-
Statistically significant discrepancies were noted in the frequencies of genotypes (CC, CT, TT) between vitiligo patients and controls (p <0.001).
-
The CC genotype predominated in controls, whereas the TT genotype was more prevalent among vitiligo patients [Table 1].
| Variable |
Cases (n=30) |
Control (n=20) |
P value | OR (95%CI) | P value |
|---|---|---|---|---|---|
| Genotype | |||||
| CC | 7 (23.4%) | 15 (75%) | X2 = 13.58 | Ref | - |
| CT | 13 (43.3%) | 4 (20%) | (df=2) | 6.9 (1.6 – 10) | 0.008* |
| TT | 10 (33.3%) | 1 (5%) | P=0.001* | 21 (2.2 – 26) | 0.007* |
| Allele | N=60 | N=40 | X2= 16.8 | ||
| C | 27 (45%) | 34 (85%) | (df=1) | Ref | |
| T | 33 (55%) | 6 (15%) | P=0.001* | 7.9 (2.8 – 12) | 0.001* |
- Chi-square test for qualitative data between groups.
*: Significant difference between groups (p-value ≤ 0.05), OR: Odds ratio, CI: Confidence interval, Ref: Reference
Distribution of TLR9 rs187084 alleles
-
There was a marked variation in allele frequencies between vitiligo patients and controls (p < 0.001), with the C allele being overrepresented in controls and the T allele exhibiting higher prevalence in vitiligo patients [Table 1].
The Hardy-Weinberg equilibrium (HWE) was assessed. The genotypic distribution was found to be in HWE, indicating a representative population sample.
Association with disease subtypes
-
The homozygous CC genotype of TLR9 (rs187084) occurred mostly in non-segmental vitiligo cases (85.7% of CC genotype instances).
-
The heterozygous CT genotype group was comprised 53.8% by cases of non-segmental vitiligo and 46.2 % by segmental vitiligo cases.
-
The TT genotype group was mostly made up of non-segmental vitiligo cases (90% of TT genotype instances) compared with 10% segmental vitiligo cases, with a non-significant p-value between the 3 genotypes (CC, CT, TT) (0.106) [Table 2].
| Variable |
CC (n=7) |
CT (n=13) |
TT (n=10) |
P value |
|---|---|---|---|---|
| #Family history | ||||
| Negative | 6 (85.7%) | 11 (84.6%) | 8 (80%) | 0.940 |
| Positive | 1 (14.3%) | 2 (15.4%) | 2 (20%) | |
| #Type | ||||
| Non-segmental | 6 (85.7%) | 7 (53.8%) | 9 (90%) | 0.106 |
| Segmental | 1 (14.3%) | 6 (46.2%) | 1 (10%) | |
| *VIDA score | ||||
| -1 | 0 (0%) | 6 (46.2%) | 4 (40%) | |
| 0 | 2 (28.6%) | 0 (0%) | 0 (0%) | |
| 1 | 0 (0%) | 1 (7.7%) | 1 (10%) | 0.110 |
| 2 | 3 (42.9%) | 4 (30.8%) | 5 (50%) | |
| 3 | 2 (28.6%) | 1 (7.7%) | 0 (0%) | |
| 4 | 0 (0%) | 1 (7.7%) | 0 (0%) | 0.251 |
| Mean ± SD | 1.75 ± 1.16 | 0.77 ± 1.8 | 0.56 ± 1.5 | |
| *Median (Interquartile) | 2 (0 – 3) | 1 (-1 - 2) | 1.5 (-1 -2) | |
| #Hair involvement | ||||
| No | 6 (85.7%) | 12 (92.3%) | 7 (70%) | 0.357 |
| Yes | 1 (14.3%) | 1 (7.7%) | 3 (30%) | |
| *Duration of disease | ||||
| Range | 1 - 25 | 1 - 15 | 2 - 37 | 0.133 |
| Mean ± SD | 6.6 ± 7.8 | 4.1 ± 3.7 | 11.3 ± 12 | |
| Median (Interquartile) | 4 (2-8) | 3 (2-5) | 4.5 (2.7 – 20) | 0.230 |
| *Onset of disease | ||||
| Early | 4 (57.1%) | 5 (38.5%) | 7 (70%) | 0.315 |
| Late | 3 (42.9%) | 8 (61.5%) | 3 (30%) |
#Fisher Exact test, SD: Standard deviation
Agarose gel electrophoresis analysis of TLR9 rs187084
-
CC genotype produced only one 145 bp band, the TT genotype produced 2 small fragments at 111 and 67 bp, and the CT genotype produced 3 bands:145, 111, and 67 bp [Figures 2 and 3].
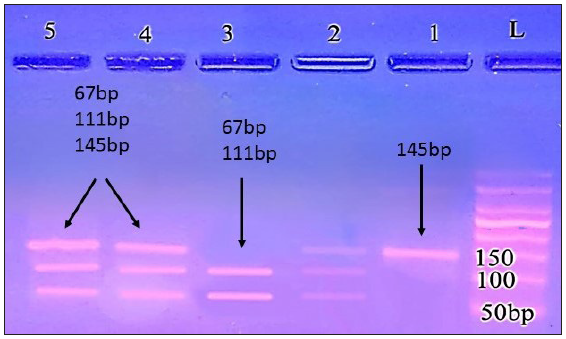
- Agarose gel electrophoresis analysis of TLR9 rs18708 in the patient group; lanes 1 (CC genotype produced only one 145 bp band); lane: 2, 5, 6 (CT genotype produced 3 bands: 145, 111 and 67 bp); lane:3 (TT genotype 111, and 67 bp bands); L: ladder 50 bp.
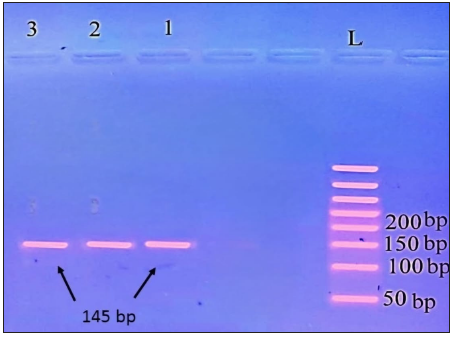
- Agarose gel electrophoresis analysis of TLR9 rs187084 in control subjects; lanes 1, 2, 3 (CC genotype produced only one 145 bp band), L: ladder 50 bp.
Association with family history
-
No significant correlation was observed between TLR9 rs187084 genotypes and a positive family history of vitiligo [Table 2].
Association with disease activity (VIDA score)
-
No statistically significant relationship emerged between TLR9 rs187084 genotypes and disease activity as assessed by the VIDA score (p=0.110) [Table 2].
Association with duration of disease
-
No significant association was established between TLR9 rs187084 genotypes and the duration of vitiligo (p=0.230) [Table 2].
Association with onset of disease
-
No statistically significant links were identified between TLR9 rs187084 genotypes and the age of onset of vitiligo (p=0.315) [Table 2].
Association with hair involvement
-
No significant association was noted between TLR9 rs187084 genotypes and hair involvement in vitiligo cases (p=0.357) [Table 2].
Genotype-phenotype association
-
The ancestral C allele of the TLR9 rs187084 polymorphism appears to confer protective effects against the development of vitiligo (p < 0.001; OR‐7.9; 95% CI‐ 2.8–12).
-
Multivariable logistic regression analysis revealed no statistically significant associations between TLR9 rs187084 genotypes and various clinical phenotypes, including gender, family history, age of onset, disease duration, type of vitiligo, and disease activity [Table 3].
| Independent variables | B | Wald X2 | OR (95% CI) | P value* |
|---|---|---|---|---|
|
Sex Male Female |
-0.1 | 0.023 |
Ref 0.9 (0.25 – 3.3) |
0.879 |
|
Family history Negative Positive |
0.223 | 0.076 |
Ref 1.924 (0.25 - 6 .1) |
0.782 |
|
Type Non-segmental Segmental |
-0.47 | 0.275 |
Ref 0.625 (0.108 – 3.6) |
0.6 |
| VIDA score | -0.401 | 3.007 | 0.670 (0.426 – 1.05) | 0.083 |
| Duration of disease | 0.028 | 0.58 | 1.03 (0.95 – 1.1) | 0.446 |
|
Onset of disease Early Late |
-0.787 | 1.83 |
Ref 0.455 (0.14 – 1.4) |
0.177 |
Discussion
The precise aetiology of vitiligo remains subject to debate, with multiple hypotheses proposed to explain its pathogenesis. Nonetheless, autoimmunity is strongly implicated in the disease’s development.16
In vitiligo, innate immunity operates via two primary signalling pathways, mediated by TLRs and nod-like receptors (NLRs), which recognize PAMPs on various microbial surfaces.4 Notably, TLR9 plays a critical role in autoimmune diseases, and current research is investigating synthetic TLR9 agonists and antagonists to regulate autoimmune inflammation.17
This study is the first to evaluate TLR9 gene polymorphisms in the context of vitiligo immunopathogenesis. Prior research has assessed TLR9 gene polymorphisms in other autoimmune and chronic inflammatory skin diseases. In our study, the CT genotype of TLR9 rs187084 was observed in 43.3% of vitiligo cases compared to 20% of controls, while the TT genotype was found in 33.3% of vitiligo cases compared to 5% of controls. These results suggest a potential genetic risk factor for vitiligo. Similarly, Bashir et al. linked the TLR9 CT and TT genotypes, particularly the T-allele, with increased susceptibility to systemic lupus erythematosus (SLE).5 This finding aligns with observations from other studies on autoimmune diseases.
Oliveira et al. found that TLR9 polymorphisms rs187084, rs5743836, and rs5743708 were associated with the development of spondyloarthritis (SpA),13 suggesting these mutations significantly impact immune response profiles and immunological regulation Zhang et al. also reported associations between TLR9 rs187084, rs5743836, and rs352162 polymorphisms and increased risk of multiple sclerosis.18 Svitich et al. suggested that TLR9 rs352140 polymorphism might increase the risk of atopic dermatitis, with the GG genotype of SNP rs352140 in TLR9 serving as a predictor for moderate AD.19Additionally, Yu et al. indicated that upregulated TLR7 and TLR9 expression might be related to melanocyte dysfunction and vitiligo pathogenesis.20
Various studies support the role of TLR gene polymorphisms in vitiligo. Traks et al. identified genetic associations between TLR gene SNPs and vitiligo susceptibility, notably TLR7 SNP 179020 and TLR3, with specific associations for TLR10 and TLR4 in subgroups.3 Karaca et al. associated TLR2 Arg753Gln and TLR4 Asp299Gly polymorphisms with vitiligo susceptibility in Turkish patients.1 Abdelsalam et al. found the protective CT genotype of TLR4 rs1927914 to be more prevalent in controls. 4 However, Rajendiran et al. did not find TLR4 rs4986790 to confer risk in the South Indian population,21 suggesting possible ethnic differences in genetic predisposition.
Our study did not find significant differences in gene polymorphism distribution with regard to family history of vitiligo, aligning with Abdelsalam et al., who found no association with TLR4 SNP rs10759932 in familial cases.4 Conversely, Traks et al. reported significant associations for TLR4 SNP rs10759932 in familial but not sporadic cases3 possibly due to differences in study size, TLRs examined, and ethnic backgrounds. Regarding disease onset, our study did not find a significant association with TLR9 gene polymorphism, unlike Karaca et al., who reported that patients with the TLR4 399IIe allele had a later onset of vitiligo.1
Our study utilised the VIDA score to assess vitiligo activity, finding no significant associations between SNPs and VIDA scores. This finding is consistent with Karaca et al., who reported no significant association between TLR4 expression and vitiligo surface area,1 and with Abdelsalam et al., who found no association between TLR4 SNP rs10759932 and disease activity.4
Limitation
The study included a smaller control group (20 individuals) compared to the case group (30 participants) due to financial constraints. To optimise resources, we prioritised including more cases to focus on the costly gene expression analysis. Due to the limited sample size, along with the methodological constraints of PCR-RFLP, there is a need for further research to validate our findings and to better understand the mechanisms through which different SNPs influence TLR9 gene function.
Conclusion
Our findings of the significant associations between CT, TT genotypes and vitiligo susceptibility underscore the crucial role of innate immunity in vitiligo pathogenesis.
Ethical approval
The research/study was approved by the Institutional Review Board at the local scientific research ethics committee at the Faculty of Medicine, Minia University, number (Approval No. 652:2023)., dated February 2023.
Declaration of patient consent
The authors certify that they have obtained all appropriate patient consent.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Use of artificial intelligence (AI)-assisted technology for manuscript preparation
The authors confirm that there was no use of artificial intelligence (AI)-assisted technology for assisting in the writing or editing of the manuscript and no images were manipulated using AI.
References
- TLR2 and TLR4 gene polymorphisms in Turkish vitiligo patients. J Eur Acad Dermatol Venereol. 2013;27:e85-90.
- [CrossRef] [PubMed] [Google Scholar]
- The Genetic Basis of Vitiligo. J Invest Dermatol. 2021;141:265-73.
- [CrossRef] [PubMed] [Google Scholar]
- Polymorphisms in toll-like receptor genes are associated with vitiligo. Front Genet. 2015;6:278.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- TLR4 gene polymorphisms in Egyptian vitiligo patients: Insights into emerging association with clinical activity, family history, and response to therapy. J Genet Eng Biotechnol. 2021;19:132.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Deleterious effect of angiotensin-converting enzyme gene polymorphism in vitiligo patients. Saudi J Biol Sci. 2021;28:4478-83.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- The genetic architecture of vitiligo. Pigment Cell Melanoma Res. 2020;33:8-15.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- the protein tyrosine phosphatase non-receptor type 22 (PTPN22) gene polymorphism and susceptibility to autoimmune diseases. In the recent topics in genetic polymorphisms. IntechOpen; 2020. Available from: http://dx.doi.org/10.5772/intechopen.90836
- Innate immune mechanisms in vitiligo: danger from within. Curr Opin Immunol. 2013;25(6):676-82.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Toll-like receptors: Role in dermatological disease. Mediators Inflamm. 2010;2010:437246.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- The association between Toll-like receptor 4 (TLR4) polymorphisms and the risk of prostate cancer in Korean men. Cancer Genet Cytogenet. 2009;190:88-92.
- [CrossRef] [PubMed] [Google Scholar]
- Association of TLR4 and TLR9 gene polymorphisms and haplotypes with cervicitis susceptibility. PLoS One. 2019;14:e0220330.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Targeting innate immunity to combat cutaneous stress: The vitiligo perspective. Front Immunol. 2021;12:613056.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Genetic polymorphisms of toll-like receptors 2 and 9 as susceptibility factors for the development of ankylosing spondylitis and psoriatic arthritis. J Immunol Res. 2019;2019:1492092.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- A single-nucleotide polymorphism-based approach for rapid and cost-effective genetic wolf monitoring in Europe based on noninvasively collected samples. Mol Ecol Resour. 2015;15:295-305.
- [CrossRef] [PubMed] [Google Scholar]
- Association of the Köbner phenomenon with disease activity and therapeutic responsiveness in vitiligo vulgaris. Arch Dermatol. 1999;135:407-13.
- [CrossRef] [PubMed] [Google Scholar]
- Vitiligo pathogenesis: Autoimmune disease, genetic defect, excessive reactive oxygen species, calcium imbalance, or what else? Exp Dermatol. 2008;17:139-40. discussion 141-60
- [CrossRef] [PubMed] [Google Scholar]
- Increased toll-like receptors activity and TLR ligands in patients with autoimmune thyroid diseases. Front Immunol. 2016;7:578.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Toll-like receptors gene polymorphisms in autoimmune disease. Front Immunol. 2021;12:672346.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Association of single nucleotide polymorphisms of TLR2, TLR4 and TLR9 with atopic dermatitis. Med. Immunol. (Russia). 2023;25:1043-8.
- [Google Scholar]
- Increased expression of Toll-like receptor 7 and 9 in vitiligo melanocytes: A pilot study. Clin Exp Dermatol. 2021;46:89-95.
- [CrossRef] [PubMed] [Google Scholar]
- Association of Nod-like receptor protein-1 (rs2670660) and Toll-like receptor-4 (rs4986790) with non-segmental vitiligo: A case-control study in South Indian population. Int J Immunogenet. 2019;46:321-30.
- [CrossRef] [PubMed] [Google Scholar]






