Translate this page into:
Malignant melanoma: Underlying epigenetic mechanisms
2 Department of Dermatology, College of Medicine, King Faisal University, Hofuf, Al Ahsa, Saudi Arabia
3 Department of Pathology, College of Medicine, Imam Abdulrahman Bin Faisal University, Dammam, Saudi Arabia
Correspondence Address:
Feroze Kaliyadan
Department of Dermatology, College of Medicine, King Faisal University, Hofuf 31982
Saudi Arabia
| How to cite this article: Sabit H, Kaliyadan F, Menezes RG. Malignant melanoma: Underlying epigenetic mechanisms. Indian J Dermatol Venereol Leprol 2020;86:475-481 |
Abstract
Although malignant melanoma is not the most common type of skin cancer, it is the most aggressive and fatal type as it can spread out and metastasize progressively. Early diagnosis and interventions lead to improved patient survival. The incidence rate of melanoma is dramatically increasing, with a few newer therapeutic options available. Therefore, establishing a reliable genetic or epigenetic-based diagnostic and prognostic tool is really important. In this review, we highlight the underlying epigenetic mechanisms involved in melanoma. Furthermore, the epigenetic-based therapeutic options will be also discussed. One of the key areas of discussion will be microRNA which is a small, single-stranded RNA molecule that serves as a regulatory element and found to regulate nearly a third of human genes. MicroRNAs play a role in a wide range of diseases including cancer. In malignant cells, it regulates cell proliferation, invasion, and metastasis.
Introduction
There were more than 18 million cancer cases diagnosed worldwide in 2018.[1] In India, melanoma is relatively rare and the commonest subtype seen clinically is the acral lentiginous type, while a mixed epithelioid and spindle cell type is histologically the most common type.[2] Cancer arises due to accumulation of genetic and/or epigenetic mutations that force cells to undergo repeated, uncontrolled divisions.[3] Skin cancers are one of the most prevalent groups of cancers—the two broad groups being melanoma and non-melanoma cancers.[4],[5],[6] Melanoma is the 19th most commonly occurring cancer in men and women, with about 300,000 new cases reported in 2018.[7] Although melanoma accounts for a smaller percentage of all skin cancers when compared to non-melanoma cancers, it is the cause for most skin cancer-related deaths.[8] Melanoma is a result of a combination of factors, including genetic and epigenetic changes.[9],[10] Several studies have revealed the role of epigenetic dysregulation in inducing melanoma. A robust association has been established between ultraviolet exposure and epigenetic alterations.[10] It has been estimated that a 10% decrease in ozone levels will result in an additional 4,500 melanoma cases worldwide every year.[11]
Genetic causes of melanoma have been extensively investigated. It is well known that melanocytes originate from the neural crest and upon differentiation, it converts to melanoblasts.[12] Activation of tumor protein p53 (P53), the genome guard, induces the biogenesis of melanosomes that help in protecting the skin.[13] With prolonged exposure to ultraviolet radiation, DNA damage results in a missense mutation in P53 which is considered an initiating event in the melanoma cascade.
Epigenetic Changes in Melanoma
Epigenetic changes can regulate gene expression without affecting the corresponding sequence of DNA.[14] These changes can affect not only DNA but also chromatin structure through posttranscriptional modifications of histone along with nucleosome remodeling. Non-coding RNAs are also implicated in epigenetic-based regulation of gene expression. These epigenetic changes play a central role in predisposition to several human diseases, including skin cancer by altering genes related to cell proliferation and apoptosis.[14],[15] A growing list of genes including, but not limited to, BRAF,[16] PTEN,[17] MGMT18 and RARB[19] are proved to be altered either genetically or epigenetically in melanoma. The fact that melanoma is controlled by a set of genes implies that treating this disease represents a challenge.
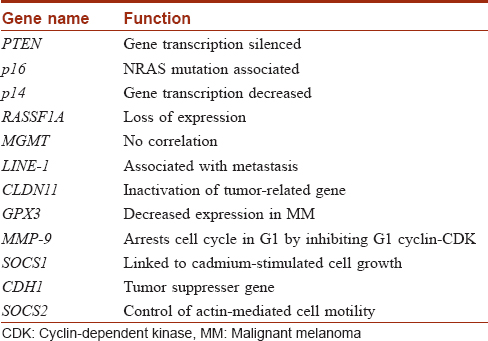
Genes hypermethylated in melanoma are presented in [Table - 1].[20],[21],[22],[23],[24],[25],[26],[27],[28],[29],[30],[31]
DNA Methylation in Melanoma
Cytosine DNA methylation is the most commonly studied epigenetic marker in the last three decades.[32] In this process, DNA methyltransferase adds a methyl group using the S-adenosyl methionine as a methyl donor. These methyl groups are added to the fifth carbon atom in Cs occurring in cytosine phosphate-guanine (CpG) dinucleotides [Figure - 1]. Methylation then recruits several proteins namely methylated DNA binding proteins (MBDs) that form a complex with histone deacetylases and chromatin remodeling factors resulting in a repressive chromatin status.[9],[33] However, cytosine methylation could be repaired by the dioxygenase family of ten-eleven translocation methylcytosine dioxygenase (TET).[34] These enzymes are unable to remove the methyl group from the cytosine residue, but instead, it hydroxylates the methyl group to form the 5-hydromethylcytosine. This can be further oxidized and finally removed by demethylation repair proteins.[35],[36] CpG dinucleotides are distributed in the human genome in a precise manner.[37] It can either occur as a dinucleotide or in clusters known as CpG islands. These islands commonly reside upstream of the gene promoter, in the first exon, where it epigenetically regulates the expression of the genes it locates within.[38],[39] Normally, CpG islands are hypomethylated or even unmethylated, but as part of neoplastic transformation, these islands become methylated, and this is a hallmark of cancer initiation and/or progression.[40] CpG island methylator phenotype status could be used to classify melanoma patients and are also found to be highly associated with mutation in ARID2 and IDH1.[41],[42] These genes are involved in chromatin remodeling. Both methylation and demethylation are securely orchestrated, where misregulation of either process results in dysregulation of cancer-related gene expression can lead to cancer development.[43] It is well known that hyper- and hypomethylation can lead to cancer, as these processes chiefly depend on the site in the genome where it takes place.[44] Generally, cancer cells undergo hypermethylation in its tumor suppressor genes along with other regulatory genes. The hypermethylation occurs mainly in the promoter regions of the specified genes, where it, upon recruiting chromatin remodeling proteins, represses the transcription process.[45] Another study has shown that dysplastic nevi are affected by promoter methylation of genes often methylated in melanoma. This is not the case with common nevi.[46] DNA hypomethylation also is found to be associated with melanoma, and this hypomethylation might lead to genomic instability. Several studies have evaluated this phenomenon. A systematic review and meta-analysis by Guo et al. found up to 50 genes, associated with risk of melanoma, in the context of promoter methylation.6 Moreover, hypomethylation-mediated inactivation of CLDN11 is reported in melanoma. Hypermethylation of tumor-related genes such as RASSF1A,[47] APC,[48] DAPK[49] HOXB13,[50] MGMT,[18] WIF1,[51] RARB,[19] INK4A,[52] SYK,[53] TFPI2[41] and SOCS154 are found to be associated with advanced melanoma and poor prognosis. In addition, the methylation status of LINE1 could serve as a prognostic biomarker for cutaneous melanoma, where patients with hypomethylation in these repeats survive better than those with hypermethylation.[55]
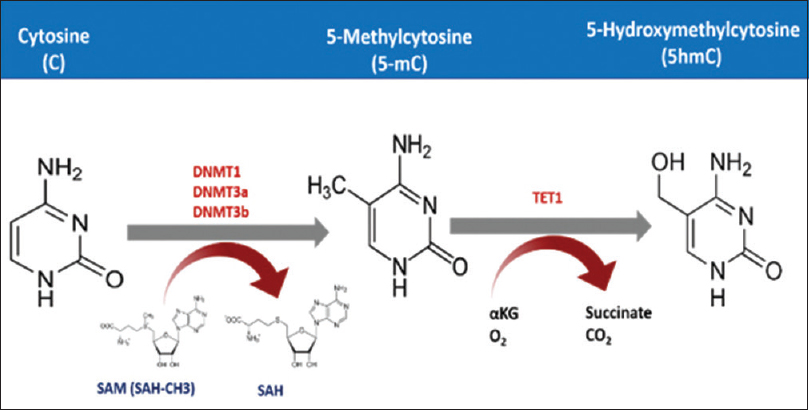 |
| Figure 1: The mechanism of cytosine methylation. DNA methyltransferase adds methyl group to the fifth carbon atom of the cytosine residue exploiting S-Adenosyl methionine as a methyl donor. TET processes this 5 mC further to finally remove the methyl group |
Specifically, it has been indicated that O-6-methylguanine-DNA methyltransferase (MGMT) promoter methylation is associated with response to dacarbazine/temozolomide (DTIC/TMZ) in disseminated cutaneous melanoma,[6] and this makes it a promising predictive marker for temozolomide therapy in a metastatic melanoma patient.[56] Meanwhile, MGMT promoter methylation may frequently coexist with P53 mutation, and these patients may benefit from treatment with alkylating agents.[57]
DNA Methytransferase (DNMT) Mutation in Melanoma
DNMTs are a group of proteins that facilitate the insertion of a methyl group to the fifth carbon atom in cytosine. DNMTs are well-characterized proteins, with specific function assigned to each member.[58] DNMT1 maintains methylation patterns in the already-methylated DNA.[59] It works mainly in the replication fork using the hemimethylated parent as a template to methylate the newly synthesized DNA strand. Whereas DNMT3A and DNMT3B are involved in the de novo methylation process in the previously unmethylated DNA.[60] Moreover, the expression of these two enzymes is shown to be associated with overall survival in stage II melanoma patients.[61] Using multivariate analysis, a study reported that the DNMT1 rs2228612 polymorphism is an independent predictor of poor overall survival in melanoma patients.[9] Disease progression is found to be an independent prognostic factor in melanoma patients.[62] Polymorphisms related to DNMT are potential targets for new therapeutic approaches. It is reported also that DNMT3B plays a protumorigenic role in human melanoma, where the lack of this protein suppresses melanoma formation in a mouse melanoma model.[63] Based on that, DNMT3 could be used as a biomarker for melanoma progression.
Histone Modifications in Melanoma
Histone is the core protein that plays a central role in terms of DNA organization in nucleosomes.[64] Nucleosomes are the basic structural unit of the chromatin structure in eukaryotic systems.[65] Histone is mainly characterized by the presence of N-rich tail areas that are rich in positively charged lysine.[66] These tails undergo epigenetic modifications that include acetylation, methylation, phosphorylation, ubiquitination and SUMO(small ubiquitin-like modifiers)-ylation.[67] These histone marks have been extensively studied, and its correlation with cancer formation is established. The most prominent histone modifications are acetylation and deacetylation, which are initiated by histone acetyltransferase and histone deacetylase proteins, respectively.[68],[69] These histone marks play a central role in the regulation of gene expression.[70] Histone acetyltransferase adds acetyl group that neutralizes the positively charged histone leading to loosening the tight binding between DNA (the negatively charged) and histone.[71] This action turns the closed heterochromatin to open euchromatin, allowing the accessibility of transcription factors and hence gene transcription. Histone deacetylase performs the opposite action, where it renders the open chromatin to a closed one, preventing the expression of the corresponding gene.[72] If these actions take place in the promoter region of cancer-related genes, the histone deacetylase activity can cause cancer to develop. Histone methylation also plays a crucial role in remodeling the chromatin and hence in the regulation of gene expression.[73] This regulation is not only depending on the methylation but also the position (at which amino acid) and degree (number of methyl groups added) of methylation.[74],[75] For example, histone H3 trimethylation at lysine 9 (K9) is generally associated with closed chromatin and hence silencing of the corresponding gene, while mono- and dimethylation of the same lysine residue is associated with open chromatin that allows the activation of the corresponding gene.[76] Furthermore, methylation of K4, K36 and K79 in H3 along with methylation of K20 in H4 is considered an active methylation tag that is associated with activated gene expression.[77] It is well established that a modified histone is associated with melanoma, where hypoacetylation causes downregulation of p21Cip1 expression along with downregulation of the proapoptotic genes such as Bim, Bak and Bax.[78] Meanwhile, histone methylation also has a role in melanoma development and progression.[79] Upregulation of Ezh2 that functions to add triple methyl groups to the K27 in H3, leads to downregulation of p21Cip1 expression in human melanoma.[80] Meanwhile, demethylating enzymes such as H3K4 demethylase JARID1B which demethylate lysine 4 at H3 have a crucial role in the development of melanoma.[81] Furthermore, lysine-specific histone demethylase demethylates histone 3 on K4 and K9 (H3K4 and H3K9).[82] It eliminates two methyl groups from H3K4me2 resulting in the formation of H3K4me1/0. This H3K4me1/0 is a modification that tags enhancers in the human genome. It is reported that enforcement of the expression of LSD1in vivo has promoted BRAFV600E-driven melanomagenesis.[83] On the other hand, downregulating LSD1 in malignant cells stimulates anti-tumor T cell immunity and inhibits cellular proliferation. H3K27 demethylase plays a vital role in transcriptional elongation and cell differentiation. Demethylating H3K27me3 leads to obliterate transcriptional repression caused by the H3K27me3 mark. UTX, the H3K27-demethylase, stimulates transcription in melanoma at sites where the promoters are tagged with trimethylated H3K27.[84] In addition, H3K4me2 is detected with high rates in melanoma samples compared to healthy skin samples. This histone mark is found to be less prevailed in metastatic melanoma compared to the primary one.
Microrna and Melanoma
MicroRNAs are endogenous, non-coding RNA transcripts of about 20-22 nucleotides in length that are evolutionarily conserved, and function as regulatory elements.[85] It is considered an epigenetic mechanism of gene expression regulation. Several types of microRNA dysregulation are found to be associated with the development and progression of melanoma [Figure - 2]. Furthermore, microRNAs could also be used as prognostic biomarkers. It has been indicated that microRNA-211 is downregulated in melanoma cell lines, 86 where the ectopic expression of this microRNA significantly inhibited its growth and invasion, suggesting that it might possess a tumor-suppressor activity.[87] Furthermore, this has been confirmed in a study that reported the location of microRNA-211 within TRPM1, a suppressor of melanoma metastasis.[88] Meanwhile, microRNA-222 is found to affect c-KIT and p27, disturbing the cell proliferation during melanoma progression. Metastasis of melanoma is further regulated by microRNA-205. MicroRNA-214 is found to interact with several tumor suppressor genes including ITGA3 and TFAP2C, where it triggers melanoma progression via suppressing these genes.[89],[90] Cancer cells survival depends on the ability to handle microenvironment components.[91] One such important factor in the tumor microenvironment is hypoxia.[92] MicroRNA-210 expression inhibits MNT and enhances cell proliferation even in the lack of oxygen.[93] MicroRNA-210 works also in the normoxic conditions, as it is upregulated in a HIF1α-dependent manner, upregulating both ATF3 and BNIP3, which function to adapt the cells to hypoxic conditions.[94],[95]
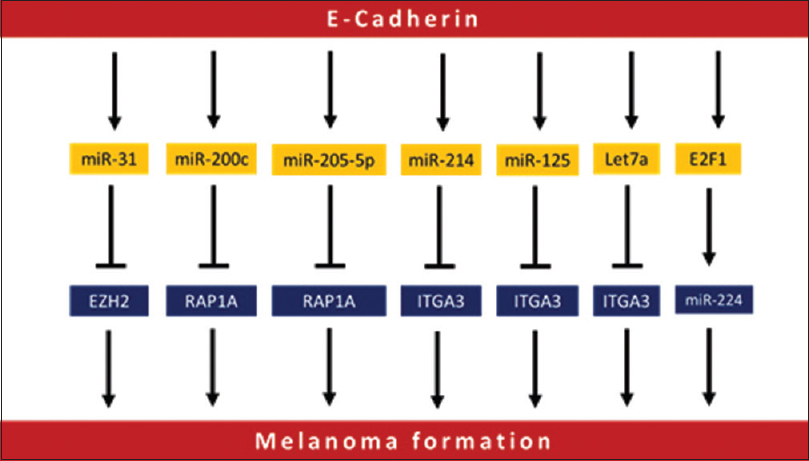 |
| Figure 2: Different microRNAs involved in melanoma initiation and progression |
In addition, microRNA-210 is detected in patients with metastatic melanoma.[96] Inversely, microRNA-33a/b and microRNA-18b are found to target HIF1α, where their expression inhibits melanoma cell proliferation.[97] Let7a has been reported to inhibit G6PD, IMPDH2, AASDHPPT, SCD and FASN, leading to induction of oxidative stress in melanoma cells.[98] Malignant cells can maneuver to avoid the hypoxic conditions via inducing angiogenesis. In melanoma, this process is found to be regulated by several microRNAs including microRNA-199a-3p, microRNA-1908 and microRNA-199a-5p.
Epigenetic-Based Therapy
The treatment of melanoma, including metastatic melanoma, has advanced in recent years with the development of newer groups such as BRAF, CTLA4 and PD1 inhibitors.[99] Notwithstanding the emergence of many new targeted therapies and immunotherapy drugs, tumor resistance to these new therapies represents a major hindrance, and hence, more efficient treatment strategies are the need of the hour. Several studies have been published in recent years to clarify the role of histone remodeling in cancer formation and progression. A wide range of histone demethylase and deacetylase inhibitors are presented as potential melanoma treatment options.[100] The recent understanding of the role of these enzymes specifies that they control the activation or repression of histone at the sites of the target genes. Histone deacetylase and histone acetyltransferase are among the proteins that were proposed as therapeutic agents for melanoma.[101] These proteins are enrolled in the clinical setting under various commercial names. For example, H3K4 and H3K9 demethylases have been used for this purpose.[102] Furthermore, JARID1 has been used in preclinical studies, and this might enhance its clinical utilization.[103] Melanoma also could be treated with mitogen-activated protein kinase inhibitors which increase the portion of JARID1B-positive cells within the whole melanoma cell population.[104] The main limitation of epigenetic-based therapies for melanoma is that they are inherently not very specific in action. It is therefore important to classify these therapies according to the specific situation.[105] Epigenetic-based interventions can help improve the sensitization of tumor cells to immunotherapy. For example, the use of histone deacetylase is associated with the upregulation of targets associated with PD1 checkpoint inhibitor therapy, which have been correlated to slower progression and better survival.[105] One of the major advantages of epigenetic-based therapies is based on the fact that epigenetic variation is more easy to be reversed as compared to genetic variations. However, the interplay between genetics and epigenetics must be explored in detail to formulate more effective treatment options.[105]
Conclusion
Epigenetic changes including histone modifications and DNA methylation play an essential role in the development of melanoma, and possibly even in its prevention and therapy. Reserving of DNA methylation status and induction of histone acetylation results in the activation of tumor-suppressor gene and silencing of oncogenes, leading to control the proliferation of melanoma cells. Studies conducted at the epigenome level have partially demonstrated the function of clusters of histone-like markers in skin cancer, although the understanding of tumors is still unclear. These studies reveal that in a particular area of DNA, histone can be more active in regulating gene expression.
Epigenetic-based therapies, although only in the initial stages of research, have potential in the treatment of melanoma, especially by increasing sensitivity of the tumor to immunotherapy. The relatively non-specific action of epigenetic-based therapies also necessitates the need to have more clarity regarding the type of treatment protocol to be used in a specific type of melanoma. More studies are needed to explore the interactions between epigenetic modifications and genetic variations in melanoma.
Furthermore, microRNAs represent a proven tool for diagnosis, prognosis, and even treatment of several types of melanoma. Clustered regularly interspaced short palindromic repeat/Cas9 is the newly emerging approach that has been employed to treat melanoma
There are some limitations regarding the use of epigenetic-based therapy for melanoma. The most important drawback of using these drugs is its off targeting that might affect normal healthy skin cells along with cancerous cells. The main challenge here is to find drugs that target the affected tissues/cells without causing unwanted effects.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
| 1. |
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424.
[Google Scholar]
|
| 2. |
Panda S, Dash S, Besra K, Samantaray S, Pathy PC, Rout N. Clinicopathological study of malignant melanoma in a regional cancer center. Indian J Cancer 2018;55:292-6.
[Google Scholar]
|
| 3. |
Wang S, Wu W. DNA methylation alterations in human cancers. In: Tollefsbol TO, editor. Epigenetics in Human Disease. 2nd ed.. Boston: Academic Press; 2019. p. 109-39.
[Google Scholar]
|
| 4. |
Verma M, Kumar V. Epigenetic drugs for cancer and precision medicine. In: Moskalev A, Vaiserman AM, editors. Epigenetics of Aging and Longevity. Boston: Academic Press; 2018. p. 439-51.
[Google Scholar]
|
| 5. |
Yang Y, Wu R, Sargsyan D, Yin R, Kuo HC, Yang I, et al. UVB drives different stages of epigenome alterations during progression of skin cancer. Cancer Lett 2019;449:20-30.
[Google Scholar]
|
| 6. |
Guo Y, Long J, Lei S. Promoter methylation as biomarkers for diagnosis of melanoma: A systematic review and meta-analysis. J Cell Physiol 2019;234:7356-67.
[Google Scholar]
|
| 7. |
Wright CY, Jean du Preez D, Millar DA, Norval M. The epidemiology of skin cancer and public health strategies for its prevention in Southern Africa. Int J Environ Res Public Health 2020;17:1017.
[Google Scholar]
|
| 8. |
Gupta AK, Bharadwaj M, Mehrotra R. Skin cancer concerns in people of color: risk factors and prevention. Asian Pac J Cancer Prev 2016;17:5257-64.
[Google Scholar]
|
| 9. |
Maric H, Supic G, Kandolf-Sekulovic L, Maric V, Mijuskovic Z, Radevic T, et al. DNMT1 and DNMT3B genetic polymorphisms affect the clinical course and outcome of melanoma patients. Melanoma Res 2019;29:596-602.
[Google Scholar]
|
| 10. |
Katiyar SK, Singh T, Prasad R, Sun Q, Vaid M. Epigenetic alterations in ultraviolet radiation-induced skin carcinogenesis: interaction of bioactive dietary components on epigenetic targets. Photochem Photobiol 2012;88:1066-74.
[Google Scholar]
|
| 11. |
Narayanan DL, Saladi RN, Fox JL. Ultraviolet radiation and skin cancer. Int J Dermatol 2010;49:978-86.
[Google Scholar]
|
| 12. |
Hu N, Strobl-Mazzulla PH, Bronner ME. Epigenetic regulation in neural crest development. Dev Biol 2014;396:159-68.
[Google Scholar]
|
| 13. |
Hsieh CC, Shen CH. The potential of targeting P53 and HSP90 overcoming acquired MAPKi-resistant melanoma. Curr Treat Options Oncol 2019;20:22.
[Google Scholar]
|
| 14. |
Lacagnina S. Epigenetics. Am J Lifestyle Med 2019;13:165-9.
[Google Scholar]
|
| 15. |
Fransquet PD, Wrigglesworth J, Woods RL, Ernst ME, Ryan J. The epigenetic clock as a predictor of disease and mortality risk: A systematic review and meta-analysis. Clin Epigenetics 2019;11:62.
[Google Scholar]
|
| 16. |
Parker D. Melanoma metastatic to brain and subsequent bleeds: The roles of BRAF status and whole brain radiotherapy. Int J Radiat Oncol Biol Phys 2019;103:40-1.
[Google Scholar]
|
| 17. |
Scortegagna M, Ruller C, Feng Y, Lazova R, Kluger H, Li JL, et al. Genetic inactivation or pharmacological inhibition of Pdk1 delays development and inhibits metastasis of Braf (V600E)::Pten(-/-) melanoma. Oncogene 2014;33:4330-9.
[Google Scholar]
|
| 18. |
Tuominen R, Jewell R, van den Oord JJ, Wolter P, Stierner U, Lindholm C, et al. MGMT promoter methylation is associated with temozolomide response and prolonged progression-free survival in disseminated cutaneous melanoma. Int J Cancer 2015;136:2844-53.
[Google Scholar]
|
| 19. |
Dahl C, Christensen C, Jönsson G, Lorentzen A, Skjødt ML, Borg Š, et al. Mutual exclusivity analysis of genetic and epigenetic drivers in melanoma identifies a link between p14 ARF and RARβ signaling. Mol Cancer Res 2013;11:1166-78.
[Google Scholar]
|
| 20. |
Lahtz C, Stranzenbach R, Fiedler E, Helmbold P, Dammann RH. Methylation of PTEN as a prognostic factor in malignant melanoma of the skin. J Invest Dermatol 2010;130:620-2.
[Google Scholar]
|
| 21. |
Jonsson A, Tuominen R, Grafström E, Hansson J, Egyhazi S. High frequency of p16(INK4A) promoter methylation in NRAS-mutated cutaneous melanoma. J Invest Dermatol 2010;130:2809-17.
[Google Scholar]
|
| 22. |
Venza M, Visalli M, Biondo C, Lentini M, Catalano T, Teti D, et al. Epigenetic regulation of p14ARF and p16INK4A expression in cutaneous and uveal melanoma. Biochim Biophys Acta 2015;1849:247-56.
[Google Scholar]
|
| 23. |
Spugnardi M, Tommasi S, Dammann R, Pfeifer GP, Hoon DS. Epigenetic inactivation of RAS association domain family protein 1 (RASSF1A) in malignant cutaneous melanoma. Cancer Res 2003;63:1639-43.
[Google Scholar]
|
| 24. |
Hoon DS. Are circulating tumor cells an independent prognostic factor in patients with high-risk melanoma? Nat Clin Pract Oncol 2004;1:74-5.
[Google Scholar]
|
| 25. |
De Araújo ÉS, Kashiwabara AY, Achatz MI, Moredo LF, De Sá BC, Duprat JP, et al. LINE-1 hypermethylation in peripheral blood of cutaneous melanoma patients is associated with metastasis. Melanoma Res 2015;25:173-7.
[Google Scholar]
|
| 26. |
Walesch SK, Richter AM, Helmbold P, Dammann RH. Claudin11 promoter hypermethylation is frequent in malignant melanoma of the skin, but uncommon in nevus cell nevi. Cancers (Basel) 2015;7:1233-43.
[Google Scholar]
|
| 27. |
Gao L, van den Hurk K, Nsengimana J, Laye JP, van den Oord JJ, Beck S, et al. Prognostic significance of promoter hypermethylation and diminished gene expression of SYNPO2 in melanoma. J Invest Dermatol 2015;135:2328-31.
[Google Scholar]
|
| 28. |
Muthusamy V, Duraisamy S, Bradbury CM, Hobbs C, Curley DP, Nelson B, et al. Epigenetic silencing of novel tumor suppressors in malignant melanoma. Cancer Res 2006;66:11187-93.
[Google Scholar]
|
| 29. |
Koga Y, Pelizzola M, Cheng E, Krauthammer M, Sznol M, Ariyan S, et al. Genome-wide screen of promoter methylation identifies novel markers in melanoma. Genome Res 2009;19:1462-70.
[Google Scholar]
|
| 30. |
Furuta J, Umebayashi Y, Miyamoto K, Kikuchi K, Otsuka F, Sugimura T, et al. Promoter methylation profiling of 30 genes in human malignant melanoma. Cancer Sci 2004;95:962-8.
[Google Scholar]
|
| 31. |
Liu S, Ren S, Howell P, Fodstad O, Riker AI. Identification of novel epigenetically modified genes in human melanoma via promoter methylation gene profiling. Pigment Cell Melanoma Res 2008;21:545-58.
[Google Scholar]
|
| 32. |
Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell 2012;150:1135-46.
[Google Scholar]
|
| 33. |
Li Y, Zhang L, Li Y, Li W, Guo Z, Li R, et al. Dynamics of DNA methylation and DNMT expression during gametogenesis and early development of scallop Patinopecten yessoensis. Mar Biotechnol (NY) 2019;21:196-205.
[Google Scholar]
|
| 34. |
Montalbán-Loro R, Lozano-Ureña A, Ito M, Krueger C, Reik W, Ferguson-Smith AC, et al. TET3 prevents terminal differentiation of adult NSCs by a non-catalytic action at Snrpn. Nat Commun 2019;10:1726.
[Google Scholar]
|
| 35. |
Kamdar S, Isserlin R, van der Kwast T, Zlotta AR, Bader GD, Fleshner NE, et al. Exploring targets of TET2-mediated methylation reprogramming as potential discriminators of prostate cancer progression. Clin Epigenetics 2019;11:54.
[Google Scholar]
|
| 36. |
Takeshima H, Ushijima T. DNA methylation changes in cancer: Mechanisms. In: Boffetta P, Hainaut P, editors. Encyclopedia of Cancer. 3rd ed.. San Diego, United States: Elsevier; 2018. p. 520-9.
[Google Scholar]
|
| 37. |
Zhang Y, Chen Y, Qu H, Wang Y. Methylation of HIF3A promoter CpG islands contributes to insulin resistance in gestational diabetes mellitus. Mol Genet Genomic Med 2019;7:e00583.
[Google Scholar]
|
| 38. |
Chai RC, Zhang KN, Liu YQ, Wu F, Zhao Z, Wang KY, et al. Combinations of four or more CpGs methylation present equivalent predictive value for MGMT expression and temozolomide therapeutic prognosis in gliomas. CNS Neurosci Ther 2019;25:314-22.
[Google Scholar]
|
| 39. |
Liao WT, You HL, Chai CY, Lee CH, Lan CE, Chang SJ, et al. Cyclin D1 promoter -56 and -54bp CpG un-methylation predicts invasive progression in arsenic-induced Bowen's disease. J Dermatol Sci 2018;89:191-7.
[Google Scholar]
|
| 40. |
Chae H, Lee S, Nephew KP, Kim S. Subtype-specific CpG island shore methylation and mutation patterns in 30 breast cancer cell lines. BMC Syst Biol 2016;10:116.
[Google Scholar]
|
| 41. |
Tanemura A, Terando AM, Sim MS, van Hoesel AQ, de Maat MF, Morton DL, et al. CpG island methylator phenotype predicts progression of malignant melanoma. Clin Cancer Res 2009;15:1801-7.
[Google Scholar]
|
| 42. |
Cancer Genome Atlas Network. Genomic classification of cutaneous melanoma. Cell 2015;161:1681-96.
[Google Scholar]
|
| 43. |
Wilting SM, Miok V, Jaspers A, Boon D, Sørgård H, Lando M, et al. Aberrant methylation-mediated silencing of microRNAs contributes to HPV-induced anchorage independence. Oncotarget 2016;7:43805-19.
[Google Scholar]
|
| 44. |
Brinkman AB, Nik-Zainal S, Simmer F, Rodríguez-González FG, Smid M, Alexandrov LB, et al. Partially methylated domains are hypervariable in breast cancer and fuel widespread CpG island hypermethylation. Nat Commun 2019;10:1749.
[Google Scholar]
|
| 45. |
Shen XF, Yuan HB, Wang GQ, Xue H, Liu YF, Zhang CX. Role of DNA hypomethylation in lateral habenular nucleus in the development of depressive-like behavior in rats. J Affect Disord 2019;252:373-81.
[Google Scholar]
|
| 46. |
Gao L, van den Hurk K, Moerkerk PTM, Goeman JJ, Beck S, Gruis NA, et al. Promoter CpG island hypermethylation in dysplastic nevus and melanoma: CLDN11 as an epigenetic biomarker for malignancy. J Invest Dermatol 2014;134:2957-66.
[Google Scholar]
|
| 47. |
Yi M, Yang J, Chen X, Li J, Li X, Wang L, et al. RASSF1A suppresses melanoma development by modulating apoptosis and cell-cycle progression. J Cell Physiol 2011;226:2360-9.
[Google Scholar]
|
| 48. |
Liu X, Li H, Wu G, Cui S. miR-182 promotes cell proliferation and invasion by inhibiting APC in melanoma. Int J Clin Exp Pathol 2018;11:1900-8.
[Google Scholar]
|
| 49. |
Rastetter M, Schagdarsurengin U, Lahtz C, Fiedler E, Marsch WCh, Dammann R, et al. Frequent intra-tumoural heterogeneity of promoter hypermethylation in malignant melanoma. Histol Histopathol 2007;22:1005-15.
[Google Scholar]
|
| 50. |
Beebe-Dimmer JL, Hathcock M, Yee C, Okoth LA, Ewing CM, Isaacs WB, et al. The HOXB13 G84E mutation is associated with an increased risk for prostate cancer and other malignancies. Cancer Epidemiol Biomarkers Prev 2015;24:1366-72.
[Google Scholar]
|
| 51. |
Park TJ, Kim M, Kim H, Park SY, Park KC, Ortonne JP, et al. Wnt inhibitory factor (WIF)-1 promotes melanogenesis in normal human melanocytes. Pigment Cell Melanoma Res 2014;27:72-81.
[Google Scholar]
|
| 52. |
Bisio A, Latorre E, Andreotti V, Bressac-de Paillerets B, Harland M, Scarra GB, et al. The 5'-untranslated region of p16INK4a melanoma tumor suppressor acts as a cellular IRES, controlling mRNA translation under hypoxia through YBX1 binding. Oncotarget 2015;6:39980-94.
[Google Scholar]
|
| 53. |
Tang H, Xu X, Xiao W, Liao Y, Xiao X, Li L, et al. Silencing of microRNA-27a facilitates autophagy and apoptosis of melanoma cells through the activation of the SYK-dependent mTOR signaling pathway. J Cell Biochem 2019;120:13262-74.
[Google Scholar]
|
| 54. |
Berzaghi R, Maia VS, Pereira FV, Melo FM, Guedes MS, Origassa CS, et al. SOCS1 favors the epithelial-mesenchymal transition in melanoma, promotes tumor progression and prevents antitumor immunity by PD-L1 expression. Sci Rep 2017;7:40585.
[Google Scholar]
|
| 55. |
Wang X, Jiang C, Fu B, Zhu R, Diao F, Xu N, et al. MILI, a PIWI family protein, inhibits melanoma cell migration through methylation of LINE1. Biochem Biophys Res Commun 2015;457:514-9.
[Google Scholar]
|
| 56. |
Tan X, Ren S, Lee W, Wu X, Rezaei K, Man YG, et al. Dual functions of miR-200b in triple-negative breast cancer metastasis and chemoimmuno-resistance. In: Proceedings of the American Association for Cancer Research Annual Meeting 2018; 2018 Apr 14-18; Chicago, IL. Philadelphia (PA): AACR. Cancer Res 2018;78 13 Suppl: Abstract No. 498.
[Google Scholar]
|
| 57. |
Yu J, Yu H, Yan J, Wu X, Yang L, Dai J, et al. Methylation of O6-methylguanine DNA methyltransferase promoter is a predictive biomarker in Chinese melanoma patients treated with alkylating agents. Transl Cancer Res 2018;7:495-505.
[Google Scholar]
|
| 58. |
Yuan Z, Chen S, Gao C, Dai Q, Zhang C, Sun Q, et al. Development of a versatile DNMT and HDAC inhibitor C02S modulating multiple cancer hallmarks for breast cancer therapy. Bioorg Chem 2019;87:200-8.
[Google Scholar]
|
| 59. |
Dasgupta H, Islam MS, Alam N, Roy A, Roychoudhury S, Panda CK. Induction of HRR genes and inhibition of DNMT1 is associated with anthracycline anti-tumor antibiotic-tolerant breast carcinoma cells. Mol Cell Biochem 2019;453:163-78.
[Google Scholar]
|
| 60. |
Liao J, Karnik R, Gu H, Ziller MJ, Clement K, Tsankov AM, et al. Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nat Genet 2015;47:469-78.
[Google Scholar]
|
| 61. |
Nguyen T, Kuo C, Nicholl MB, Sim MS, Turner RR, Morton DL, et al. Downregulation of microRNA-29c is associated with hypermethylation of tumor-related genes and disease outcome in cutaneous melanoma. Epigenetics 2011;6:388-94.
[Google Scholar]
|
| 62. |
Elder DE. Melanoma progression. Pathology 2016;48:147-54.
[Google Scholar]
|
| 63. |
Micevic G, Muthusamy V, Damsky W, Theodosakis N, Liu X, Meeth K, et al. DNMT3b Modulates melanoma growth by controlling levels of mTORC2 component RICTOR. Cell Rep 2016;14:2180-92.
[Google Scholar]
|
| 64. |
Longbotham JE, Chio CM, Dharmarajan V, Trnka MJ, Torres IO, Goswami D, et al. Histone H3 binding to the PHD1 domain of histone demethylase KDM5A enables active site remodeling. Nat Commun 2019;10:94.
[Google Scholar]
|
| 65. |
Maldonado R, Schwartz U, Silberhorn E, Längst G. Nucleosomes stabilize ssRNA-dsDNA triple helices in human cells. Mol Cell 2019; 73:1243-1254.e6
[Google Scholar]
|
| 66. |
Chan JC, Maze I. Histone crotonylation makes its mark in depression research. Biol Psychiatry 2019;85:616-8.
[Google Scholar]
|
| 67. |
Borgermann N, Ackermann L, Schwertman P, Hendriks IA, Thijssen K, Liu JC, et al. SUMOylation promotes protective responses to DNA-protein crosslinks. EMBO J 2019;38:e101496.
[Google Scholar]
|
| 68. |
Zhao C, Gao J, Zhang L, Su L, Luan Y. Novel HDAC6 selective inhibitors with 4-aminopiperidine-1- carboxamide as the core structure enhanced growth inhibitory activity of bortezomib in MCF-7 cells. Biosci Trends 2019;13:91-7.
[Google Scholar]
|
| 69. |
Icardi L, De Bosscher K, Tavernier J. The HAT/HDAC interplay: multilevel control of STAT signaling. Cytokine Growth Factor Rev 2012;23:283-91.
[Google Scholar]
|
| 70. |
Scholz B, Schulte JS, Hamer S, Himmler K, Pluteanu F, Seidl MD, et al. HDAC (histone deacetylase) inhibitor valproic acid attenuates atrial remodeling and delays the onset of atrial fibrillation in mice. Circ Arrhythm Electrophysiol 2019;12:e007071.
[Google Scholar]
|
| 71. |
Xu S, Hui Y, Shu J, Qian J, Li L. Characterization of the human mucin 5AC promoter and its regulation by the histone acetyltransferase P300. Int J Mol Med 2019;43:1263-70.
[Google Scholar]
|
| 72. |
Roche J, Bertrand P. Inside HDACs with more selective HDAC inhibitors. Eur J Med Chem 2016;121:451-83.
[Google Scholar]
|
| 73. |
Chory EJ, Calarco JP, Hathaway NA, Bell O, Neel DS, Crabtree GR. Nucleosome turnover regulates histone methylation patterns over the genome. Mol Cell 2019;61-72.e3.
[Google Scholar]
|
| 74. |
Greer EL, Shi Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat Rev Genet 2012;13:343-57.
[Google Scholar]
|
| 75. |
Sun H, Zhou X, Chen H, Li Q, Costa M. Modulation of histone methylation and MLH1 gene silencing by hexavalent chromium. Toxicol Appl Pharmacol 2009;237:258-66.
[Google Scholar]
|
| 76. |
Rondinelli B, Gogola E, Yücel H, Duarte AA, van de Ven M, van der Sluijs R, et al. EZH2 promotes degradation of stalled replication forks by recruiting MUS81 through histone H3 trimethylation. Nat Cell Biol 2017;19:1371-8.
[Google Scholar]
|
| 77. |
Sein H, Värv S, Kristjuhan A. Distribution and maintenance of histone H3 lysine 36 trimethylation in transcribed locus. PLoS One 2015;10:e0120200.
[Google Scholar]
|
| 78. |
Saha K, Hornyak TJ, Eckert RL. Epigenetic cancer prevention mechanisms in skin cancer. AAPS J 2013;15:1064-71.
[Google Scholar]
|
| 79. |
Orouji E, Utikal J. Tackling malignant melanoma epigenetically: Histone lysine methylation. Clin Epigenetics 2018;10:145.
[Google Scholar]
|
| 80. |
Zingg D, Debbache J, Schaefer SM, Tuncer E, Frommel SC, Cheng P, et al. The epigenetic modifier EZH2 controls melanoma growth and metastasis through silencing of distinct tumour suppressors. Nat Commun 2015;6:6051.
[Google Scholar]
|
| 81. |
Kuźbicki Ł, Lange D, Stanek-Widera A, Chwirot BW. Prognostic significance of RBP2-H1 variant of JARID1B in melanoma. BMC Cancer 2017;17:854.
[Google Scholar]
|
| 82. |
Yang RF, Zhao GW, Liang ST, Chen HZ, Liu DP. Lysine-specific demethylase 1 represses THP-1 monocyte-to-macrophage differentiation. Chin Med Sci J 2013;28:82-7.
[Google Scholar]
|
| 83. |
Yu Y, Schleich K, Yue B, Ji S, Lohneis P, Kemper K, et al. Targeting the senescence-overriding cooperative activity of structurally unrelated H3K9 demethylases in melanoma. Cancer Cell 2018;33:322-36.e8.
[Google Scholar]
|
| 84. |
Le Guellec S, Macagno N, Velasco V, Lamant L, Lae M, Filleron T, et al. Loss of H3K27 trimethylation is not suitable for distinguishing malignant peripheral nerve sheath tumor from melanoma: A study of 387 cases including mimicking lesions. Mod Pathol 2017;30:1677-87.
[Google Scholar]
|
| 85. |
Salinas-Vera YM, Marchat LA, Gallardo-Rincón D, Ruiz-García E, Astudillo-De La Vega H, Echavarría-Zepeda R, et al. AngiomiRs: microRNAs driving angiogenesis in cancer (review). Int J Mol Med 2019;43:657-70.
[Google Scholar]
|
| 86. |
Bell RE, Khaled M, Netanely D, Schubert S, Golan T, Buxbaum A, et al. Transcription factor/microRNA axis blocks melanoma invasion program by miR-211 targeting NUAK1. J Invest Dermatol 2014;134:441-51.
[Google Scholar]
|
| 87. |
Xu Y, Brenn T, Brown ER, Doherty V, Melton DW. Differential expression of microRNAs during melanoma progression: MiR-200c, miR-205 and miR-211 are downregulated in melanoma and act as tumour suppressors. Br J Cancer 2012;106:553-61.
[Google Scholar]
|
| 88. |
Mazar J, DeYoung K, Khaitan D, Meister E, Almodovar A, Goydos J, et al. The regulation of miRNA-211 expression and its role in melanoma cell invasiveness. PLoS One 2010;5:e13779.
[Google Scholar]
|
| 89. |
Dettori D, Orso F, Penna E, Salmena L, Pandolfi PP, Caselle M, et al. miR-214 in stroma cells and tumor progression. Eur J Cancer 2016;61:S94.
[Google Scholar]
|
| 90. |
Penna E, Orso F, Cimino D, Tenaglia E, Lembo A, Quaglino E, et al. microRNA-214 contributes to melanoma tumour progression through suppression of TFAP2C. EMBO J 2011;30:1990-2007.
[Google Scholar]
|
| 91. |
Dougan SK. The pancreatic cancer microenvironment. Cancer J 2017;23:321-5.
[Google Scholar]
|
| 92. |
Attri KS, Mehla K, Singh PK. Evaluation of Macrophage Polarization in Pancreatic Cancer Microenvironment under Hypoxia. Methods in Molecular Biology. New York: Springer; 2018. p. 265-76.
[Google Scholar]
|
| 93. |
Sun H, Zhang Z, Grandori C. Abstract A65: MicroRNA miR-210 Modulates Cellular Response to Hypoxia through the MYC Antagonist MNT. Cell Cycle Regulators. American Association for Cancer Research; 2009.
[Google Scholar]
|
| 94. |
Martinez VD, Firmino NS, Marshall EA, Ng KW, Wadsworth BJ, Anderson C, et al. Non-coding RNAs predict recurrence-free survival of patients with hypoxic tumours. Sci Rep 2018;8:152.
[Google Scholar]
|
| 95. |
Bavelloni A, Ramazzotti G, Poli A, Piazzi M, Focaccia E, Blalock W, et al. MiRNA-210: A current overview. Anticancer Res 2017;37:6511-21.
[Google Scholar]
|
| 96. |
Grosso S, Doyen J, Parks SK, Bertero T, Paye A, Cardinaud B, et al. MiR-210 promotes a hypoxic phenotype and increases radioresistance in human lung cancer cell lines. Cell Death Dis 2013;4:e544.
[Google Scholar]
|
| 97. |
Romano G, Kwong LN. miRNAs, Melanoma and Microenvironment: An Intricate Network. Int J Mol Sci 2017; 18 (11). pii: E2354
[Google Scholar]
|
| 98. |
Serguienko A, Grad I, Wennerstrøm AB, Meza-Zepeda LA, Thiede B, Stratford EW, et al. Metabolic reprogramming of metastatic breast cancer and melanoma by let-7a microRNA. Oncotarget 2015;6:2451-65.
[Google Scholar]
|
| 99. |
Davis LE, Shalin SC, Tackett AJ. Current state of melanoma diagnosis and treatment. Cancer Biol Ther 2019;20:1366-79.
[Google Scholar]
|
| 100. |
Swahari V, West AE. Histone demethylases in neuronal differentiation, plasticity, and disease. Curr Opin Neurobiol 2019;59:9-15.
[Google Scholar]
|
| 101. |
Thambyrajah R, Fadlullah MZH, Proffitt M, Patel R, Cowley SM, Kouskoff V, et al. HDAC1 and HDAC2 modulate TGF-β signaling during endothelial-to-hematopoietic transition. Stem Cell Reports 2018;10:1369-83.
[Google Scholar]
|
| 102. |
Liu Y, Qin S, Chen TY, Lei M, Dhar SS, Ho JC, et al. Structural insights into trans-histone regulation of H3K4 methylation by unique histone H4 binding of MLL3/4. Nat Commun 2019;10:36.
[Google Scholar]
|
| 103. |
Mocavini I, Pippa S, Licursi V, Paci P, Trisciuoglio D, Mannironi C, et al. JARID1B expression and its function in DNA damage repair are tightly regulated by miRNAs in breast cancer. Cancer Sci 2019;110:1232-43.
[Google Scholar]
|
| 104. |
Zhou AY, Johnson DB. Combinatorial therapies in melanoma: MAPK inhibitors and beyond. Am J Clin Dermatol 2018;19:181-93.
[Google Scholar]
|
| 105. |
Moran B, Silva R, Perry AS, Gallagher WM. Epigenetics of malignant melanoma. Semin Cancer Biol 2018;51:80-8.
[Google Scholar]
|
Fulltext Views
7,550
PDF downloads
2,891





