Translate this page into:
Association between exonic polymorphisms of human leukocyte antigen-G gene and non-segmental vitiligo in the Korean population
Su-Kang Kim and Hyo-Eun Kwon have contributed equally to this work.
*Corresponding author: Ki-Heon Jeong, Department of Dermatology, College of Medicine, Kyung Hee University, Seoul, South Korea. mdfamily@naver.com
-
Received: ,
Accepted: ,
How to cite this article: Kim SK, Kwon HE, Jeong KH, Shin MK, Lee MH. Association between exonic polymorphisms of human leukocyte antigen-G gene and non-segmental vitiligo in the Korean population. Indian J Dermatol Venereol Leprol 2022;88:749-54.
Abstract
Background
Vitiligo is a pigmentary skin disorder characterised by a chronic and progressive loss of melanocytes. Although several theories have been suggested to the pathogenesis of vitiligo, an autoimmune process leading to melanocyte destruction appears most likely. Human leukocyte antigen-G is a non-classic, major histocompatibility complex Class I molecule that plays an important role in the suppression of the immune response. Several recent studies have provided evidences that polymorphisms in the human leukocyte antigen-G gene might be related with autoimmune diseases.
Objectives
The aim of this study was to decide whether exonic single nucleotide polymorphisms in human leukocyte antigen-G contribute to the risk of developing non-segmental vitiligo in the Korean population.
Methods
To evaluate the associations between exonic single nucleotide polymorphisms (rs1630223 [Ala5Ala] and rs12722477 [Leu134Ile]) of human leukocyte antigen-G and vitiligo, 244 patients with vitiligo and 398 healthy controls were recruited. Genotyping was performed using Fluidigm 192.24 Dynamic Array with EP1 (Fluidigm Corp., CA). The SNP type assay (Fluidigm Corp., CA), which employs allele-specifically designed fluorescences (FAM or VIC) primers and a common reverse primer was applied and the data were analysed using the EP1 single nucleotide polymorphisms genotyping analysis software to obtain genotype calls.
Results
Two exonic single nucleotide polymorphisms (rs1630223 and rs12722477) exhibited significant associations with susceptibility and remained a statistically significant association following Bonferroni correction. These two single nucleotide polymorphisms were located within a block of linkage disequilibrium. Haplotypes G-C and A-A comprising rs1630223 and rs12722477 demonstrated a significant association with non-segmental vitiligo.
Limitations
The protein expression level of patients with vitiligo and controls was not studied and a replication study of the genetic association in an independent group was not managed.
Conclusion
Our results suggest that exonic human leukocyte antigen-G polymorphisms (rs1630223 and rs12722477) are associated with the development of non-segmental vitiligo.
Keywords
Genome-wide association study
human leukocyte antigen-G antigens
polymorphism
vitiligo
Plain Language Summary
Although the aetiology of vitiligo is still unknown, several theories have been proposed to explain the pathogenesis of vitiligo. Among them, genetic factors are also known to play key roles. More than 40 susceptible loci have been identified to be associated with vitiligo. In this study, we conducted an association study of 244 vitiligo patients and 398 controls of the Korean population. The results suggest that exonic single nucleotide polymorphisms (rs1630223 and rs12722477) of human leukocyte antigen-G have a significant association with the development of vitiligo in this population.
Introduction
Vitiligo is an acquired pigmentary skin disorder in which melanocytes are destroyed by an autoimmune process, leading to a hypopigmented zone in the affected skin tissue.1 It can be divided into two clinical subtypes: Segmental and non-segmental vitiligo. Segmental vitiligo appears in a dermatomal distribution while the non-segmental varieties include generalised, acrofacial, or mixed types.2
Non-segmental vitiligo is considered as an autoimmune disease because it is often associated with other autoimmune diseases such as rheumatoid arthritis, adult-onset insulin-dependent diabetes mellitus, autoimmune thyroid disease, systemic lupus erythematous and Addison’s disease in patients and their close relatives.3,4 Therefore, considering its autoimmunity and relationship to other autoimmune disorders, genetic associations of innate immunity have been investigated.1
Innate immune responses may drive the destruction of melanocytes in cellular stress, such as interferon-γ-STAT1-CXCL10 signaling.5,6 However, these pathways may be affected by various inflammatory responses, including hepatitis, airway epithelium, or systemic lupus erythematosus.7-10 It has been found that many genes in the genomic region are involved in vitiligo development in genome-wide studies such as CTLA4, PTPN22, RERE, TYR and CASP7.1,11
The human major histocompatibility complex class is located on chromosome 6p21 and contains several loci, most of which encode immune regulatory molecules. The presentation of the major histocompatibility complex Class I antigens on antigen-presenting cells to cytotoxic T lymphocytes plays an important role in the protection of the body against intracellular pathogens.12 Human leukocyte antigen-G was first described by Geraghty et al. in 1987 and is a non-classical major histocompatibility complex molecule, which also includes human leukocyte antigen-E and F.13 It was initially described as restricted to extravillous trophoblasts, the foetal cell population directly in contact with maternal tissues, providing inhibitory signals to both natural killer (NK) cells and T cells, presumably helping to maintain maternofetal tolerance.14 However, recently, the human leukocyte antigen-G protein was found in the cornea, thymic medulla, mesenchymal stem cells and endothelial precursors in healthy, non-foetal subjects.15-18
Human leukocyte antigen-G has eight exons and seven introns and its sequence is approximately 86% identical to the consensus sequence of the human leukocyte antigen-A, B and C genes. Alternative splicing of the primary transcript generates has seven isoforms, including four membrane-bound (human leukocyte antigen-G1 to human leukocyte antigen-G4) isoforms and three secreted soluble isoforms (human leukocyte antigen-G5 to human leukocyte antigen-G7).13 HLA-G can suppress almost all phases in the immune response. Its expression is restricted to some tissues under normal conditions, but increases strongly in pathological conditions such as melanoma, pemphigus vulgaris and gastrointestinal cancer.19-21
Although the mechanism for these associations between the human leukocyte antigen system and vitiligo is unknown, Jalel et al. reported a significant negative correlation between human leukocyte antigen-G expression and vitiligo among Tunisians.22 In our earlier study, we observed a distinct association between human leukocyte antigen -DQB1 promoter polymorphism (rs1736936) of human leukocyte antigen-G, human leukocyte antigen -G 14 bp insertion/deletion polymorphism and nonsegmental vitiligo in the Korean population.23,24 Based on these results, the objective of this study was to investigate whether the exonic polymorphisms of human leukocyte antigen-G contribute to the risk of developing non-segmental vitiligo in the Korean population.
Material and Methods
Study subjects
Patients with non-segmental vitiligo who visited department of dermatology Kyung Hee University, Seoul, South Korea were enrolled. Healthy subjects were recruited following a general health check-up programme. All subjects were unrelated Korean individuals. All patients aged over 18 years with either active or stable bilateral vitiligo lesions were included in the study. The subjects were excluded if they were pregnant or breastfeeding. Patients with segmental vitiligo were also excluded from the study. A total of 244 patients with non-segmental vitiligo (127 males and 117 females with an average age of 40.9 years) and 398 healthy subjects (210 males and 188 females with an average age of 53.8 years) were enrolled [Table 1]. Active nonsegmental vitiligo characterised by dissemination of existing lesions and/or appearance of new lesions within the previous six months were detected in 166 patients, whereas 78 patients showed stable disease (no increase in lesion size or number within six months). A family history of non-segmental vitiligo was elicited in 38 patients. Nine patients had concomitant autoimmune disease (autoimmune thyroiditis, diabetes mellitus Type 1, or systemic lupus erythematous). Control subjects had no symptoms or disease of relevance to the study. Informed consent was obtained from all participants. This study was approved by the Institutional Review Board of Kyung Hee University Hospital (Institutional Review Board number: 2015-07-111) and was in accordance with the Helsinki guidelines.
| NSV | Control | |
|---|---|---|
| Number (male/female), n | 244 (127/117) | 398 (210/188) |
| Age (mean±SD, years) | 40.9±18.0 | 53.8±17.1 |
| Disease activity | ||
| Active | 166 | |
| Stable | 78 | |
| Family history of NSV, n | ||
| (+) | 38 | |
| (–) | 206 | |
| Presence of autoimmune disease* in NSV, n | ||
| (+) | 9 | |
| (–) | 235 |
Single nucleotide polymorphism selection and genotyping
Based on the International HapMap Project dataset (http://www.hapmap.org ), the human leukocyte antigen-G single nucleotide polymorphisms with the optimal minor allele frequency (>0.10) and the best coverage to serve as tag single nucleotide polymorphisms for four ethnic groups (Caucasians in Utah, Han Chinese in Beijing, Japanese in Tokyo and Yoruba in Ibadan, Nigeria) were selected. Among the single nucleotide polymorphisms, previously researched single nucleotide polymorphisms that were associated with several diseases, including vitiligo, were prioritised and two exonic single nucleotide polymorphisms were selected. The single nucleotide polymorphisms comprised rs1630223 (Ala5Ala) and rs12722477 (Leu134Ile).
Genomic DNA was extracted from peripheral blood using a blood genomic DNA isolation kit (Roche, Indianapolis, IN, USA). Each single nucleotide polymorphism was genotyped using Fluidigm 192.24 Dynamic Array integrated fluid circuits (Fluidigm Incorporated, San Francisco, CA, USA). First, for the polymerase chain reaction analysis, the amount of DNA was quantified to 50 ng and the DNA fragment was amplified using ×1 Qiagen multiplex polymerase chain reaction master mix (Qiagen, PN 206143). The integrated fluid circuits and EP-1 systems were used to perform polymerase chain reaction. The integrated fluid circuits controller was used to automatically supply DNA and reagents. After amplification by polymerase chain reaction, images were obtained for genotyping in the EP-1 system. Genotyping was determined by analysing the data using the Fluidigm single nucleotide polymorphism genotyping software.
In silico analysis for single nucleotide polymorphisms
To evaluate that – single nucleotide polymorphisms could cause amino acid changes, in silico analysis was performed using SIFT dbSNP (http://genetics.bwh.harvard.edu/pph2/index.shtml ) and PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/ ).
Statistical analysis
First, the Hardy-Weinberg equilibrium was tested in the control group using a Chi-square test. To analyse the relationship between each single nucleotide polymorphisms and susceptibility to nonsegmental vitiligo, logistic regression models were evaluated. The models were codominant1, codominant2, dominant, recessive and log-additive. odds ratios (OR), 95% confidence intervals (CI) and P-values were determined. All statistical analyses were performed using SNPStats (http://bioinfo.iconcologia.net/index.php?module=Snpstats ). Haplotype analysis among single nucleotide polymorphisms was performed using haploview 4.2. The difference in haplotype frequency was tested using a Chi-square test and logistic regression. The significance level for statistical tests was set at P < 0.05.
Results
Genotypic and allelic frequencies in patients with non-segmental vitiligo and unaffected controls
Hardy-Weinberg equilibrium for genotype distributions of two single nucleotide polymorphisms in the control group (rs1630223, P = 0.31 and rs12722477, P = 0.75) and in the non-segmental vitiligo group (rs1630223, P = 0.009 and rs12722477, P = 0.03) was shown. The distributions of genotypic and allelic frequencies of each single nucleotide polymorphisms are shown in Table 2. The distribution of A/A, A/G and G/G genotypes in rs1630223 single nucleotide polymorphisms was 147 (37.0%), 197 (49.6%) and 53 (13.3%) in the control group and 39 (16%), 151 (61.9%) and 54 (22.1%) in the non-segmental vitiligo group, respectively. There was a significant association between genotype frequency and distribution in the non-segmental vitiligo group in the dominant, recessive and log-addictive models (OR = 2.79, 95%, CI = 1.84–4.22, P < 0.001 in the dominant model; OR = 1.61, 95%, CI = 1.03–2.50, P = 0.037 in the recessive model; and OR = 1.83, 95%, CI = 1.41–2.39, P < 0.001 in the log-additive model). This association remained significant, following Bonferroni correction in the dominant model (Pc < 0.001) and log-addictive model (Pc < 0.001). The G allele frequency of rs1630223 was higher in the non-segmental vitiligo group (53.1%) than in the control group (38.2%). The difference was significantly associated with non-segmental vitiligo risk (OR = 1.83, 95% CI = 1.46–2.30, P < 0.001, Pc < 0.001).
| SNPs | Genotype/allele | Control n (%) | NSV n (%) | Models | OR (95% CI) | P-value | P-valuec |
|---|---|---|---|---|---|---|---|
| rs1630223 | A/A | 147(37.0) | 39(16.0) | Dominant | 2.79(1.84–4.22) | <0.001 | <0.001 |
| Ala10Ala | A/G | 197(49.6) | 151(61.9) | Recessive | 1.61(1.03–2.50) | 0.037 | 0.74 |
| G/G | 53(13.3) | 54(22.1) | Log-additive | 1.83(1.41–2.39) | <0.001 | <0.001 | |
| A | 491(61.8) | 229(46.9) | |||||
| G | 303(38.2) | 259(53.1) | 1.83(1.46–2.30) | <0.001 | <0.001 | ||
| rs12722477 | C/C | 160(40.2) | 129(53.5) | Dominant | 0.61(0.43–0.85) | 0.004 | 0.008 |
| Leu134Ile | A/C | 187(47.0) | 103(42.7) | Recessive | 0.29(0.14–0.61) | <0.001 | <0.001 |
| A/A | 51(12.8) | 9(3.7) | Log-additive | 0.59(0.45–0.78) | <0.001 | <0.001 | |
| C | 507(63.7) | 361(74.9) | |||||
| A | 289(36.3) | 121(25.1) | 0.59(0.46–0.76) | <0.001 | <0.001 |
Bold numbers indicate significant association. Missing genotype data were omitted for accurate analysis. HLA-G: Human leukocyte antigen G, SNP: Single nucleotide polymorphism, NSV: Non-segmental vitiligo, n: Number of subjects, OR: Odds ratio, CI: Confidence interval, Pc: Corrected P value
The distributions of C/C, A/C and A/A genotypes in rs12722477 single nucleotide polymorphisms were 160 (40.2%), 187 (47.0%) and 51 (12.8%) in the control group and 129 (53.5%), 103 (42.7%) and nine (3.7%) in the nonsegmental vitiligo group, respectively. There were significant associations between genotype frequency and distribution in the non-segmental vitiligo group in the dominant, recessive and log-addictive models (OR = 0.61, 95% CI = 0.43–0.85, P = 0.004 in the dominant model; OR = 0.29, 95% CI = 0.14–0.61, P = 0.003 in the recessive model; and OR = 0.59, 95%, CI = 0.45–0.78, P < 0.001 in the log-additive model). This association remained significant, following Bonferroni correction in the dominant model (Pc = 0.008), recessive model (Pc < 0.001) and log-addictive model (Pc < 0.001). The allele frequency of rs12722477 was lower in the nonsegmental vitiligo group (25.1%) than in the control group (36.3%). The difference was significantly associated with non-segmental vitiligo risk (OR = 0.59, 95%, CI = 0.46–0.76, P < 0.001, Pc < 0.001).
Association between Human leukocyte antigen-G haplotypes and non-segmental vitiligo
To further analyse the haplotype structure, we characterised the linkage disequilibrium between the human leukocyte antigen-G single nucleotide polymorphisms in control subjects using pair-wise Ds (data not shown). The two single nucleotide polymorphisms (rs1630223 and rs12722477) that showed significant association with non-segmental vitiligo were located within a block of linkage disequilibrium. Haploview 4.2 was used to further analyse haplotypes among the two examined single nucleotide polymorphisms. Three haplotypes were detected (haplotype G-C, frequency = 0.438; A-A, frequency = 0.321 and A-C, frequency = 0.241). Haplotypes G-C and A-A were associated with non-segmental vitiligo (haplotype G-C, P < 0.001; haplotype A-A, P < 0.001) [Table 3].
| Block | Haplotype frequency | Vitiligo group | Control group | Chi-square | P-value | ||
|---|---|---|---|---|---|---|---|
| + | – | + | – | ||||
| GC | 0.438 | 259.0 | 229.0 | 303.6 | 492.4 | 27.38 | <0.001 |
| AA | 0.321 | 122.7 | 365.3 | 289.0 | 507.0 | 17.30 | <0.001 |
| AC | 0.241 | 106.3 | 381.7 | 203.4 | 592.6 | 2.35 | 0.13 |
A haplotype with a frequency of more than 0.1 is shown. The haplotypes comprised rs1630223 and rs12722477. Bold numbers indicate significant association. HLA-G: Human leukocyte antigen G, SNP: Single nucleotide polymorphism, NSV: Non-segmental vitiligo
In silico analysis for single nucleotide polymorphisms
Since non-synonymous single nucleotide polymorphisms could cause amino acid changes, in silico analysis was performed for the two single nucleotide polymorphisms (rs1630223 and rs12722477). In SIFT, rs1630223 was a synonymous single nucleotide polymorphisms and amino acid did not change, resulting in “tolerated.” rs12722477 was a non-synonymous single nucleotide polymorphisms and the result was “deleterious” in SIFT and “probably damaging (score 0.995)” in Polyphen-2 analysis.
Association between human leukocyte antigen-G polymorphisms and non-segmental vitiligo parameters
We investigated the differences between the polymorphisms according to the clinical parameters of non-segmental vitiligo. There were no significant differences in genotype or haplotype frequencies for polymorphisms based on age and gender, disease activity and the presence of a positive family history.
Discussion
Unlike segmental vitiligo, non-segmental vitiligo provides evidence of a major autoimmune component, including the occurrence of autoantibodies against melanin in the affected individuals.25 Non-segmental vitiligo is polygenic in its inheritance. Alleles at many unlinked loci are associated with increased disease susceptibility.26 In recent years, linkage analysis and candidate gene studies have indicated various genes as affecting susceptibility to non-segmental vitiligo as well as being involved in disease pathogenesis.27 These include the immunoregulatory genes, human leukocyte antigen genes, apoptotic and cytotoxic genes, melanocyte-related genes and susceptible genes with unknown functions.28 However, the exact genetic defects leading to non-segmental vitiligo have not been clarified. Complex genetic susceptibility may contribute to non-segmental vitiligo, with candidate genes related in epidermal physiology and self-destruction of melanocytes.29
In depigmented lesions of vitiligo patients, cellular infiltrates are accompanied with progressive loss of melanocytes. It is postulated that these cellular infiltrates, especially T cells, are actively cytolytic against the residual melanocytes. This autoimmune response may progress further in the absence of functional regulatory T cells.30 A decrease in the number of regulatory T cells and the activity of NK cells are observed in the skin of patients with autoimmune diseases including vitiligo.31 Human leukocyte antigen-G expression has been suggested to be involved in these mechanisms.
Human leukocyte antigen-G plays a variety of roles including proliferation, differentiation, cytokine secretion, cytolysis and immunoglobulin production, such as inducing apoptosis of activated CD8+ T cells, interacting with T regulatory cells, modulating the activity of NK cells and dendritic cells and blocking of the allo-cytotoxic T lymphocyte.32-36 Various polymorphisms of human leukocyte antigen-G generate polymorphic protein chains that bind and present peptide antigens to T lymphocytes.37 The coding region of human leukocyte antigen-G reveals a few polymorphic sites randomly distributed along exons and introns, contrary to the high frequency of exonic polymorphisms observed in classical human leukocyte antigen Class I genes. Recently, several studies have suggested that human leukocyte antigen-G polymorphisms are associated with autoimmune diseases such as systemic lupus erythematous and juvenile idiopathic arthritis.38,39
In this study, we investigated the potential influence of exonic human leukocyte antigen-G polymorphisms on the development of non-segmental vitiligo in the Korean population. Our data suggests that two single nucleotide polymorphisms, rs1630223 and rs12722477 are significantly associated with non-segmental vitiligo. In addition, we examined linkage disequilibrium between the single nucleotide polymorphisms and conducted a haplotype analysis between patients with non-segmental vitiligo and control subjects. Single nucleotide polymorphisms rs1630223 and rs12722477 are located within a block of strong linkage disequilibrium. Haplotype analysis demonstrated that the haplotypes G-C and A-A are significantly associated with non-segmental vitiligo. In silico analysis, rs12722477 was “deleterious” in SIFT and “probably damaging” (score 0.995) in Polyphen-2 and this result could be expected that rs12722477 affects protein structure changes and functions.
Our results suggest that human leukocyte antigen-G polymorphisms may have a potential function in the pathogenesis of non-segmental vitiligo. The exact mechanism by which this pathogenesis is brought about remains unknown; however, the reduction of cutaneous expression of human leukocyte antigen-G in the skin of patients with vitiligo compared to healthy controls has been reported in several studies.22 Taken together, these findings indicate that polymorphisms in human leukocyte antigen-G may have influence on the expression of human leukocyte antigen-G in non-segmental vitiligo lesions, resulting in the activation of inflammatory responses of CD8+ T cells and dendritic cells. Furthermore, the cascade of the reaction leads to the suppression of regulatory T cells and the destruction of melanocytes [Figure 1]. To better understand the pathogenesis, further functional studies are required.
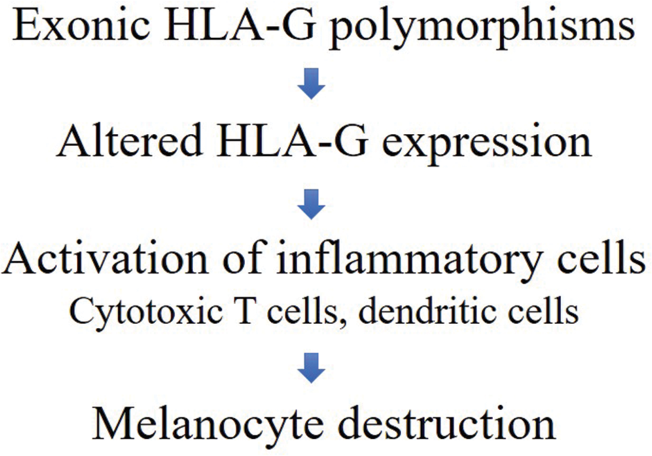
- The proposed mechanism of the relationship between HLA-G polymorphisms and NSV, HLA-G: Human leukocyte antigen G, NSV: Nonsegmental vitiligo
The limitations of this study are: a) although the study investigated the single nucleotide polymorphisms of human leukocyte antigen-G, the protein expression level in the skin or blood samples of patients with vitiligo and controls was not studied and b) a replication study of the genetic association in an independent group was not managed.
Conclusion
We conducted a genetic association analysis of the exonic polymorphism of human leukocyte antigen-G and the risk of development of non-segmental vitiligo. Our results suggest that rs1630223 and rs12722477 are involved in the pathogenesis of non-segmental vitiligo. Further studies are needed to investigate their precise role.
Declaration of patient consent
Institutional Review Board (IRB) permission obtained for the study.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- Genome-wide association studies of autoimmune vitiligo identify 23 new risk loci and highlight key pathways and regulatory variants. Nat Genet. 2016;48:1418-24.
- [CrossRef] [PubMed] [Google Scholar]
- The definition and assessment of vitiligo: A consensus report of the vitiligo European task force. Pigment Cell Res. 2007;20:27-35.
- [CrossRef] [PubMed] [Google Scholar]
- Epidemiology of vitiligo and associated autoimmune diseases in Caucasian probands and their families. Pigment Cell Res. 2003;16:208-14.
- [CrossRef] [PubMed] [Google Scholar]
- Early disease onset and increased risk of other autoimmune diseases in familial generalized vitiligo. Pigment Cell Res. 2005;18:300-5.
- [CrossRef] [PubMed] [Google Scholar]
- Recent advances in understanding vitiligo. F1000Res. 2016;5:F1000. Faculty Rev-2234
- [CrossRef] [PubMed] [Google Scholar]
- Understanding mechanisms of autoimmunity through translational research in vitiligo. Curr Opin Immunol. 2016;43:81-8.
- [CrossRef] [PubMed] [Google Scholar]
- Interleukin-27 and IFNγ regulate the expression of CXCL9, CXCL10, and CXCL11 in hepatitis. J Mol Med (Berl). 2015;93:1355-67.
- [CrossRef] [PubMed] [Google Scholar]
- Isoliquiritigenin inhibits interferon-γ-inducible genes expression in hepatocytes through down-regulating activation of JAK1/STAT1, IRF3/MyD88, ERK/ MAPK, JNK/MAPK and PI3K/Akt signalling pathways. Cell Physiol Biochem. 2015;37:501-14.
- [CrossRef] [PubMed] [Google Scholar]
- IFN-γ-induced JAK/STAT, but not NF-κB, signalling pathway is insensitive to glucocorticoid in airway epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2015;309:L348-59.
- [CrossRef] [PubMed] [Google Scholar]
- Positive correlation of STAT1 and miR-146a with anaemia in patients with systemic lupus erythematosus. J Clin Immunol. 2014;34:171-80.
- [CrossRef] [PubMed] [Google Scholar]
- Six decades of vitiligo genetics: Genome-wide studies provide insights into autoimmune pathogenesis. J Invest Dermatol. 2012;13:268-73.
- [CrossRef] [PubMed] [Google Scholar]
- Antigen processing and presentation by the class I major histocompatibility complex. Annu Rev Immunol. 1996;14:369-96.
- [CrossRef] [PubMed] [Google Scholar]
- A human major histocompatibility complex Class I gene that encodes a protein with a shortened cytoplasmic segment. Proc Natl Acad Sci USA. 1987;84:9145-9.
- [CrossRef] [PubMed] [Google Scholar]
- Immunomodulatory properties of HLA-G in infectious diseases. J Immunol Res. 2014;2014:298569.
- [CrossRef] [PubMed] [Google Scholar]
- Expression of HLA-G in human cornea, an immune-privileged tissue. Hum Immunol. 2003;64:1039-44.
- [CrossRef] [PubMed] [Google Scholar]
- Primary cultured human thymic epithelial cells express both membrane-bound and soluble HLA-G translated products. J Reprod Immunol. 1999;43:225-34.
- [CrossRef] [Google Scholar]
- Human leukocyte antigen-G5 secretion by human mesenchymal stem cells is required to suppress T lymphocyte and natural killer function and to induce CD4+CD25highFOXP3+ regulatory T cells. Stem Cells. 2008;26:212-22.
- [CrossRef] [PubMed] [Google Scholar]
- Erythroblasts secrete the nonclassical HLA-G molecule from primitive to definitive hematopoiesis. Blood. 2004;104:3153-60.
- [CrossRef] [PubMed] [Google Scholar]
- Analysis of HLA antigen expression in benign and malignant melanocytic lesions reveals that upregulation of HLA-G expression correlates with malignant transformation, high inflammatory infiltration and HLA-A1 genotype. Int J Cancer. 2004;108:243-50.
- [CrossRef] [PubMed] [Google Scholar]
- HLA-G is associated with pemphigus vulgaris in Jewish patients. Hum Immunol. 2004;65:39-46.
- [CrossRef] [PubMed] [Google Scholar]
- HLA-G expression in tumour tissues and soluble HLA-G plasma levels in patients with gastrointestinal cancer. Asian Pac J Cancer Prev. 2018;19:2731-5.
- [Google Scholar]
- Impact of HLA-G in the outcome of vitiligo in Tunisian patients. Indian J Dermatol. 2010;55:25-8.
- [CrossRef] [PubMed] [Google Scholar]
- HLADQB1 gene polymorphisms and non-segmental vitiligo: A case-control study in the Korean population. J Dermatol Sci. 2016;82:48-50.
- [CrossRef] [PubMed] [Google Scholar]
- Association between an HLA-G 14 bp insertion/deletion polymorphism and non-segmental vitiligo in the Korean population. Arch Dermatol Res. 2014;306:577-82.
- [CrossRef] [PubMed] [Google Scholar]
- Therapeutic implications of autoimmune vitiligo T cells. Autoimmun Rev. 2006;5:486-92.
- [CrossRef] [PubMed] [Google Scholar]
- Genetic epidemiology of vitiligo: A study of 815 probands and their families from south China. Int J Dermatol. 2006;45:1176-81.
- [CrossRef] [PubMed] [Google Scholar]
- The genetics of generalized vitiligo. Curr Dir Autoimmun. 2008;10:244-57.
- [CrossRef] [PubMed] [Google Scholar]
- Genetic susceptibility to vitiligo: GWAS approaches for identifying vitiligo susceptibility genes and loci. Front Genet. 2016;7:3.
- [CrossRef] [PubMed] [Google Scholar]
- Presence of T cells and macrophages in inflammatory vitiligo skin parallels melanocyte disappearance. Am J Pathol. 1996;148:1219-28.
- [Google Scholar]
- Imbalance of regulatory T cells in human autoimmune diseases. Immunology. 2006;117:289-300.
- [CrossRef] [PubMed] [Google Scholar]
- Cutting edge: Soluble HLA-G1 triggers CD95/CD95 ligand-mediated apoptosis in activated CD8+ cells by interacting with CD8. J Immunol. 2000;164:6100-4.
- [CrossRef] [PubMed] [Google Scholar]
- Human leukocyte antigen-G is closely associated with tumour immune escape in gastric cancer by increasing local regulatory T cells. Cancer Sci. 2011;102:1272-80.
- [CrossRef] [PubMed] [Google Scholar]
- A soluble HLA-G protein that inhibits natural killer cell-mediated cytotoxicity. Transplant Proc. 2001;33:2355-9.
- [CrossRef] [Google Scholar]
- Modulation of dendritic cell differentiation by HLA-G and ILT4 requires the IL-6--STAT3 signaling pathway. Proc Natl Acad Sci USA. 2008;105:8357-62.
- [CrossRef] [PubMed] [Google Scholar]
- HLA-G has a concentration-dependent effect on the generation of an allo-CTL response. Immunology. 2000;101:191-200.
- [CrossRef] [PubMed] [Google Scholar]
- Association of HLA alleles and haplotypes with vitiligo in Moroccan patients: A case-control study. Arch Dermatol Res. 2013;305:925-32.
- [CrossRef] [PubMed] [Google Scholar]
- HLA-G genotype and HLA-G expression in systemic lupus erythematosus: HLA-G as a putative susceptibility gene in systemic lupus erythematosus. Tissue Antigens. 2008;71:520-9.
- [CrossRef] [PubMed] [Google Scholar]
- Association of the HLA-G 14-bp insertion/deletion polymorphism with juvenile idiopathic arthritis and rheumatoid arthritis. Tissue Antigens. 2008;71:440-6.
- [CrossRef] [PubMed] [Google Scholar]






