Translate this page into:
Association between PITX2 polymorphism and androgenetic alopecia in the Indian population
Corresponding author: Dr. Ravikumar Sambandam, Department of Medical Biotechnology, Aarupadai Veedu Medical College & Hospital, Vinayaka Mission’s Research Foundation (Deemed to be University) Kirumampakkam, Puducherry, India. ravikumar.sambandam@avmc.edu.in
-
Received: ,
Accepted: ,
How to cite this article: Murugan M, Sadasivam IP, Manoharan A, Jayakumar S, Vetriselvan Y, Samuel MS, et al. Association between PITX2 polymorphism and androgenetic alopecia in the Indian population. Indian J Dermatol Venereol Leprol. 2025;91:158-62. doi: 10.25259/IJDVL_1147_2023
Abstract
Background
Androgenetic alopecia, also known as male pattern baldness, is a common form of hair loss influenced by environmental, hormonal, and genetic factors. According to recent research, the PITX2 gene may play a key role in the pathophysiology of androgenetic alopecia (AGA).
Objectives
This study examines the association between genetic variants of the PITX2 gene and AGA risk.
Methods
The genomic DNA was extracted from peripheral blood samples collected from 70 male AGA patients and 60 non-androgenetic alopecia controls. The isolated DNA was quantified and the genotype for three PITX2 polymorphisms (rs2200733, rs10033464, and rs13143308) was identified using TaqMan assays. The statistical analysis was done to determine the allele frequency of genetic variants between AGA and non-AGA groups.
Results
The demographic profile of the study population showed that the AGA and non-AGA groups differed in age. The AGA group had higher blood pressure, a higher prevalence of smoking, alcohol consumption, metabolic syndrome, insulin resistance, and a higher incidence of family history. Through genetic analysis, significant correlations were found between AGA risk and specific PITX2 polymorphisms, significantly with the rs2200733 allele (OR = 6.08, p < 0.001*), the rs1003464 G allele (OR = 2.02, p < 0.019*) and the rs13143308 showed GT genotype (OR = 4.26, p < 0.001*).
Conclusion
Based on our findings, the PITX2 polymorphisms may play a vital role in the development of AGA. This study also found the interactions between genetic and environmental factors in AGA pathogenesis.
Keywords
PITX2
Androgenetic alopecia
Single nucleotide polymorphism
Genetic variation.
Introduction
Androgenetic alopecia (AGA), or male pattern baldness, results from genetic, hormonal, and environmental factors affecting hair follicles. This widespread dermatological condition affects 85% of men and 40% of women globally.1-4 In India, the prevalence rate among males aged 30–50 years is high as 58%.5 AGA is often accompanied by comorbidities such as metabolic syndrome, endocrine disorders, cardiovascular diseases, hypertension.6 insulin resistance,7 abnormal lipid profiles,8 obesity, smoking,9 and mental health issues. Recent research findings demonstrate a statistically significant correlation between androgenetic alopecia (AGA) and coronary artery diseases10 particularly among younger patients when compared to a control group.11 Previous studies have indicated that AGA patients may be at increased risk of developing cardiovascular diseases12 and metabolic syndrome13 with a higher risk observed in those experiencing severe balding14
PITX2 is a homeobox gene that encodes for a transcription factor PITX2 which is present in chromosome 4. PITX2 is involved in the pituitary, eye, tooth, and skeletal muscle formation, particularly in the left-right asymmetry of the heart, and other developmental processes.15-17 Several diseases, including appendicitis, atrial fibrillation18, and Axenfeld-Rieger syndrome,19 have been linked to genetic variants in or near the PITX2 gene. The PITX2 contributes to AGA by regulating specific genes via the Wnt/ß-catenin signalling pathway, which is essential for the growth of hair follicles.20 One important mediator of AGA is the androgen receptor (AR), whose expression may be regulated by PITX2. The hair follicle growth cycle is modulated by AR through its binding to dihydrotestosterone (DHT), a potent androgen derived from testosterone. PITX2 may also have an impact on 5-alpha reductase activity which is the enzyme responsible for converting testosterone to DHT. To clarify the molecular mechanisms underlying androgenetic alopecia, additional research is necessary as the precise role of PITX2 in this condition. In the patients with AGA, levels of PITX2 expression were significantly lower than in non-balding subjects.21 Downregulating PITX2 may result in increased AR activity and hair loss because it may function as a negative regulator of AR expression. Comparing androgenetic alopecia patients’ hair follicles to those of healthy controls revealed an increase in PITX2 expression.4 According to these studies, PITX2 may function as a positive regulator of 5-alpha reductase expression, and hair loss and increased DHT production may be the consequences of its overexpression. According to a genome-wide association study, men who have a variant of PITX2 are more likely to experience androgenetic alopecia. PITX2 was found to be expressed less in balding scalps than in non-balding scalps and it was found to control the expression of genes linked to the hair growth cycle. As a result, PITX2 could be a promising target for the treatment of androgenetic alopecia because it regulates the expression which promotes hair growth rather than inhibits it. By understanding PITX2 role in androgenetic alopecia, which leads to the development of personalised gene-based therapies. Our research attempts to explore the possible relationship between androgenetic alopecia and genetic variations in the 4q25 region of the PITX2 gene. The results of the current study could improve our knowledge in understanding of the genetic architecture of androgenetic alopecia.
Methods
Subject selection and DNA isolation
In this case-control study, we recruited 70 androgenetic alopecia patients and 60 non-androgenetic controls from a tertiary care hospital in Puducherry, India. The Institutional Human Ethical Committee of our institute approved the study with reference number AVMC/IEC2019/63. All the participants signed their consent forms after clarifications with the research assistant. The 2 mL of blood samples were collected from study participants in Ethylene Diamine Tetra-Acetic acid (EDTA) -coated blood collection tubes. Genomic DNA was isolated from 200 µl of blood sample using QIAamp DNA Blood Mini Kit (Qiagen, Germany) followed by manufacture protocol. Isolated DNA samples were stored at −20℃ until use. The DNA bands were checked by using Agarose gel electrophoresis and quantified by using Nanodrop.
Genotyping of PITX2 gene SNPs
The Single nucleotide polymorphism (SNP) rs2200733, rs10033464, and rs13143308 genotyping was performed using a TaqMan assay kit (Thermo Scientific) in Quant studio 5 Real-Time PCR System. Briefly, 1 μl of 50 ng of genomic DNA was added with 5 μl of 2× master mix, 0.25 μl of 20× SNP assay mix, and 3.75 μl of nuclease-free water to make a final volume of 10 μl. The thermal cycler program was set as 10 min at 95℃, 40 cycles of 20 s at 95℃, and 30 s at 60℃.
Data analyses
The genotyping was done in Quant Studio 5 Real-Time PCR Systems with Quant Studio design and analysis software v1.5.2. Data analysis was carried out by Microsoft Excel version 19 and SPSS version 28 version. Data are presented as frequencies and percentages. The chi-square test was used to find any significant association between qualitative variables. A p-value of 0.05 was taken as the margin for establishing statistical significance. The diagrammatic flow chart of the workflow is shown in Figure 1.
- Flow chart shows the workflow of methodology.
Results
Baseline characteristics of the study population
The demographics of the study population included 70 subjects with androgenetic alopecia (AA) and 60 controls with non-androgenetic alopecia (NAA). The mean age of the androgenetic alopecia group was 49.83 years (±15.3), slightly higher than the mean age of the non-androgenetic alopecia group which was 43.15 years (±14.6). The androgenetic alopecia group had a slightly higher mean BMI of 26.10 kg/m2 (±3.6), compared to the non-androgenetic alopecia group’s mean of 24.6 kg/m2 (±3.9). In addition, the androgenetic alopecia group exhibited greater systolic and diastolic blood pressures than the non- AGA group. Other clinical parameters, including fasting blood sugar, fasting serum insulin, triglycerides, and high-density lipoproteins, showed no statistically significant differences between the two groups. There was a significant difference in family history, alcohol intake, and metabolic syndrome between the two groups (p values <0.01, and <0.05, respectively). Variables associated with smoking and insulin resistance showed no significant differences between the groups [Table 1].
| Variables | Case (n = 70) | Control (n = 60) | P value |
|---|---|---|---|
| Age (years) | 49.83±15.3 | 43.15±14.6 | <0.05* |
| Sex (male/female) | 70/0 | 60/0 | |
| Body Mass Index (kg/m2) | 26.10±3.6 | 24.6±3.9 | <0.05* |
| SBP | 124.5±10.3 | 119.8±9.6 | <0.01* |
| DBP | 84.7±9.2 | 81.43±9.12 | <0.05* |
| Fasting blood sugar | 116.3±36.7 | 113.4±40.4 | 0.668 |
| Fasting serum insulin | 8.5±6.5 | 8.5±4.2 | 1.000 |
| Triglycerides | 149.6±47.1 | 174.2±241.9 | 0.406 |
| High-density lipoproteins | 45.8±7.9 | 46.2±9.1 | 0.788 |
| Family history (yes/no) | 47/23 | 19/41 | <0.01* |
| Alcohol intake (yes/no) | 38/32 | 18/42 | <0.05* |
| Smoking (yes/no) | 28/42 | 21/39 | 0.557 |
| Metabolic syndrome (yes/no) | 31/39 | 14/46 | <0.05* |
| Insulin resistance (yes/no) | 24/46 | 15/45 | 0.249 |
The variables compared included age, sex, BMI: Body mass index, family history, alcohol intake, smoking, metabolic syndrome, and insulin resistance for 70 androgenetic alopecia/60 non-androgenetic alopecia individuals. (SBP: Systolic blood pressure, DBP: Diastolic blood pressure, TG: Triglycerides, HDL: High-density lipoproteins, FBS: Fasting blood sugar.) The * denotes the significant p-value less than 0.05.
Genotyping association of SNPs in PITX2 with AGA
The genetic analysis of three PITX2 polymorphisms (rs2200733, rs1003464, and rs13143308) in AGA patients and non- AGA controls were shown in Tables 2, 3, and 4. For rs2200733, the CT genotype was significantly associated with (OR = 6.08, p < 0.001*) and the T allele also showed a significant association (OR = 3.24, p < 0.001*) [Table 2]. In the case of rs1003464, the TG and GG genotypes did not show significant associations individually, while the combination of T/G + G/G genotypes had a borderline significant association (OR = 2.01, p = 0.060), as did the G allele (OR = 2.02, p < 0.019*) [Table 3]. Finally, rs13143308, the GT genotype was significantly associated with AA (OR = 4.26, p < 0.001*), as was the G allele (OR = 0.41, p < 0.01*) [Table 4]. The graphical representation of the genotype distribution of rs2200733, rs10033464, and rs13143308 of the PITX2 gene is shown in Figure 2.
| Genotype | Cases (n = 70) | Control (n = 60) | OR (95% CI) | P value |
|---|---|---|---|---|
| CC | 30 (43%) | 48 (80%) | Reference | |
| CT | 38 (54%) | 10 (17%) | 6.08 (2.64–13.98) | <0.001* |
| TT | 2 (3%) | 2 (3%) | 1.6 (0.21–11.97) | 0.644 |
| Dominant Model | ||||
| C/C | 30 (43%) | 48 (80%) | Reference | |
| C/T + T/T | 40 (57%) | 12 (20%) | 5.33 (2.42–11.75) | < 0.001* |
| Recessive Model | ||||
| C/C+C/T | 68 (97%) | 58 (97%) | Reference | |
| T/T | 2 (3%) | 2 (3%) | 0.85 (0.12–6.25) | 0.875 |
| Alleles | ||||
| C | 98 (70%) | 106 (88%) | Reference | |
| T | 42 (30%) | 14 (12%) | 3.24 (1.67–6.31) | < 0.001* |
The table represents the comparison of rs2200733 genotype frequencies between the 70 androgenetic alopecia/60 non-androgenetic alopecia group, along with odds ratios (OR) and p values. The * denotes the significant p-value less than 0.01. (CI: Confidence interval.)
| Genotype | Cases (n = 70) | Control (n = 60) | OR (95% CI) | P value |
|---|---|---|---|---|
| TT | 39 (55%) | 43 (71%) | Reference | |
| TG | 20 (29%) | 13 (22%) | 1.7 (0.75–3.86) | 0.205 |
| GG | 11 (16%) | 4 (7%) | 3.03 (0.89–10.31) | 0. 066 |
| Dominant Model | ||||
| T/T | 39 (56%) | 43 (80%) | Reference | |
| T/G + G/G | 31 (44%) | 17 (20%) | 2.01 (0.97–4.19) | 0.060 |
| Recessive Model | ||||
| T/T+T/G | 69 (97%) | 56 (93%) | Reference | |
| G/G | 11 (3%) | 4 (7%) | 2.23 (0.67–7.39) | 0.179 |
| Alleles | ||||
| T | 98 (70%) | 99 (82%) | Reference | |
| G | 42 (30%) | 21 (18%) | 2.02 (1.12–3.66) | <0.019* |
The table represents the comparison of rs1003464 genotype frequencies between the 70 androgenetic alopecia/60 non-androgenetic alopecia group, along with odds ratios (OR) and p values. The asterisks indicate statistically significant differences (cases versus controls) as determined by Chi-Square tests: p-value < 0.05. (CI: Confidence interval.)
| Genotype | Cases (n = 70) | Control (n = 60) | OR (95% CI) | P value |
|---|---|---|---|---|
| GG | 25 (36%) | 41 (68%) | Reference | |
| GT | 39 (55%) | 15 (25%) | 4.26 (1.96–9.26) | <0.001* |
| TT | 6 (AQ269%) | 4 (7%) | 2.46 (0.63–9.58) | 0.437 |
| Dominant Model | ||||
| G/G | 25(36%) | 41 (68%) | Reference | |
| G/T + T/T | 45 (64%) | 19 (32%) | 3.88 (1.87–8.07) | <0.001* |
| Recessive Model | ||||
| G/G+G/T | 64 (91%) | 56 (93%) | Reference | |
| T/T | 6 (9%) | 4 (7%) | 1.31 (0.35–4.89) | 0.684 |
| Alleles | ||||
| A | 89 (64%) | 97 (81%) | Reference | |
| G | 51 (36%) | 23 (19%) | 0.41 (0.23–0.73) | <0.01* |
The table represents the comparison of rs13143308 genotype frequencies between the 70 androgenetic alopecia/60 non-androgenetic alopecia group, along with odds ratios (OR) and p values. The asterisks indicate statistically significant differences (cases versus controls) as determined by Chi-Square tests: p-value < 0.05. (CI: Confidence interval.)
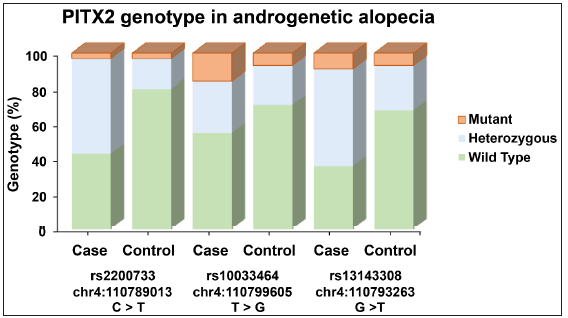
- Graphical representation of the genotype distribution of rs2200733, rs10033464, and rs13143308 of the PITX2 gene. The y-axis denotes the percentage of genotype and the x-axis denote the number of cases and controls in each genotype variant of the bar graph in androgenetic alopecia patients and controls.
SNP–SNP interactions among rs2200733, rs10033464 and rs10033464 and their association with AGA
The analysis of SNP–SNP reveals interactions among three genetic variants (rs2200733, rs10033464, and rs10033464) and their association with androgenetic alopecia (AGA) patients and controls. To investigate the interaction among rs2200733 (C to T substitution, risk allele = T), rs10033464 (T to G substitution, risk allele = G), and rs13143308 (G to T substitution, risk allele = T), the analysis is based on the number of risk alleles individuals possesses. It compares the distribution of these risk alleles between AGA cases and controls. We used a non-risk allele as a baseline or reference and estimated the OR for 1, 2, 3, 4, 5, and 6 risk alleles [Table 5]. The findings reveal a strong and statistically significant association between the number of risk alleles and the likelihood of being an AGA case. Notably, individuals with one risk allele are 13.22 times more likely to have AGA than controls and this association is highly significant (p = 0.0003). Similarly, individuals with two, three, or four risk alleles also exhibit significantly elevated odds of being AGA cases (p < 0.01). The 95% confidence intervals for all odds ratios do not include 1, further emphasising the robustness of these associations.
| Number of Risk alleles | Cases (n = 70) | Controls (n = 60) | OR (95% CI) | P value |
|---|---|---|---|---|
| 0 | 12 (17%) | 34 (57%) | Reference | |
| 1 | 14 (20%) | 3 (5%) | 13.22 (3.2–54.1) | 0.0003* |
| 2 | 21 (30%) | 17 (28%) | 3.50 (1.3–8.7) | 0.0075* |
| 3 | 13 (19%) | 3 (5%) | 12.27 (2.97–50.6) | 0.0005* |
| 4 | 10 (14%) | 3 (5%) | 9.4 (2.21–40.19) | 0.0024* |
The table represents the SNP–SNP interactions among rs2200733, rs10033464, and rs13143308 genotypes between the 70 androgenetic alopecia/60 non-androgenetic alopecia group, along with odds ratios (OR) and p values. The * denotes the significant p-value less than 0.05. (CI: Confidence interval.)
Discussion
The findings from this study reflect the complicated interplay between genetic factors and environment in connection to androgenetic alopecia and also demonstrate a challenging research question regarding the precise genetic mechanism involved. The study’s demographic profile shows some of the differences between the AGA group and the non-AGA group. Furthermore, the AGA group provided higher average systolic and diastolic blood pressure which may be related to a link between AGA with hypertension. This supports previous studies that have linked vascular factors as contributors to hair loss in men. In addition, the AGA group had more people with a family history of alopecia, metabolic syndrome, smoking, insulin resistance, and alcohol intake which highlights its multifactorial nature. The genetic makeup of AGA as well as lifestyle choices like smoking or drinking or even metabolic disorders might cause its emergence and further development.22
In this study, we have found a connection between hair loss in men and the genetic variation of the PITX2 gene. The role of the PITX2 gene involves the development of embryos especially concerning the left-right asymmetry of the heart.23 PITX2 has been identified as the candidate gene associated with Axenfeld-Rieger syndrome which is an autosomal dominant hereditary condition manifested by ocular, craniofacial, and umbilical malformation as well as occasional aberrations in the limb, pituitary gland, or heart among others.24 Various studies have shown that β-catenin controls the PITX2 expression in different systems. An increase in the Wnt/Dvl/β-catenin signaling pathway can result in activation levels of cyclin D2 and c-myc by raising expressions of PITX2 genes which are specifically involved in cardiac outflow tract development.25 The second instance indicates β-catenin-dependent downstream activity targeting PITX2 where atrial separation during cardiac morphogenesis is a critical step.26 In follicular differentiation in-vitro systems, positive regulation of β-catenin activity occurs via the action of PITX2 factors on outer root sheath cells.27
In this study, the genetic analysis of AGA risk associated with PITX2 polymorphisms (rs2200733, rs1003464, and rs13143308) was studied. Our study found that a strong association between the AGA patient and controls in rs2200733 polymorphism with CT genotype [OR = 6.08; 95% CI = 2.64– 3.98; p = < 0.001*] along with a significant association for the T allele [OR = 3.24; 95% CI = 1.67–6.31; p = < 0.001*]. The rs13143308 polymorphism with GT genotype of [OR = 4.26; 95% CI = 1.96–9.26; p = < 0.001*] and G allele [OR = 0.41; 95% CI = 0.23–0.73; p = < 0.01*] shows a significant association. The rs10033464 SNP was significantly associated with G allele [OR = 2.02; 95% CI = 1.12 – 3.66 P = < 0.019*]. Thus, these results would provide the PITX2 gene may also be one of the important genes in AGA pathogenesis because as changes occur in this genetic region, it may lead to abnormal functioning of androgenetic-dependent mechanisms which are necessary for normal hair follicle functioning. Susceptibility to AGA may be influenced by a genetic variation that has the potential to impact the PITX2 gene. The role of the PITX2 gene in the cycle of hair follicle development is significant. The development of mature hair follicles involves three distinct stages: anagen, catagen, and telogen.
Conclusion
These findings suggest that PITX2 polymorphisms may contribute to the development of AGA. Furthermore, this research revealed the complex interplay of genetic and environmental factors in AGA pathogenesis.
Androgens play a significant role in modulating hair cycle activities by targeting their specific receptors on the dermal papilla cells or other surrounding components of hair follicle structures, thus causing premature development of terminal hairs in pre-adolescence. This dysregulation may lead to several common hair disorders such as androgenetic alopecia. The advancement in this field widens the therapeutic interventions with personalised treatment approaches. The elucidation of PITX2’s involvement in androgenetic alopecia serves as a model of complex relationships between genetic factors and hair biology that enables the development of new solutions to deal with this common problem. This research examines the correlation between the intergenic region of chromosome 4q25 upstream from the PITX2 gene and AGA that contributes to proving the mutation in this locus among AGA patients. In people with AGA, frequently occurring genetic variants at locus 4q25, namely, rs2200733, rs13143308, and rs10033464 have indicated an undeniable statistical relationship. To the best of our knowledge, this is the first study to demonstrate a significant correlation between rs2200733 and rs13143308 within the PITX2 gene in AGA cases in the Indian population. The limitation of the study is the relatively small sample size, which means our findings need to be validated by larger studies.
Ethical approval
The research/study was approved by the Institutional Review Board at Aarupadai Veedu Medical College & Hospital, number AVMC/IEC2019/63, dated 30.11.2019.
Declaration of patient consent
The authors certify that they have obtained all appropriate patient consent.
Financial support and sponsorship
The first author sincerely acknowledges the ICMR Senior Research Fellowship Program (3/1//1(25)/2022-NCD-1) for financial support. Additionally, this study received financial support from the ICMR Adhoc Project No.5/4-35 5/1-6/2020-NCD-II Project.
Conflicts of interest
There are no conflicts of interest.
Use of artificial intelligence (AI)-assisted technology for manuscript preparation
The authors confirm that there was no use of artificial intelligence (AI)-assisted technology for assisting in the writing or editing of the manuscript and no images were manipulated using AI.
References
- Is early onset androgenic alopecia a marker of metabolic syndrome and carotid artery atherosclerosis in young indian male patients? Int J Trichology. 2015;7:141-7.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Male androgenetic alopecia: A population-based study in 1,005 subjects. Int J Trichology. 2009;1:131-3.
- [CrossRef] [PubMed] [Google Scholar]
- Genetic and molecular aspects of androgenetic alopecia. Indian J Dermatol Venereol Leprol. 2018;84:263-8.
- [CrossRef] [PubMed] [Google Scholar]
- Comorbidities in androgenetic alopecia: A comprehensive review. Dermatol Ther (Heidelb). 2022;12:2233-47.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Androgenetic alopecia: An update. Indian J Dermatol Venereol Leprol. 2013;79:613-25.
- [CrossRef] [PubMed] [Google Scholar]
- Association of androgenetic alopecia and hypertension. Eur J Dermatol. 2007;17:220-2.
- [CrossRef] [PubMed] [Google Scholar]
- Early androgenetic alopecia as a marker of insulin resistance. Lancet. 2000;356:1165-6.
- [CrossRef] [PubMed] [Google Scholar]
- Evaluation of lipid levels in androgenetic alopecia in comparison with control group. J Eur Acad Dermatol Venereol. 2009;23:80-1.
- [CrossRef] [PubMed] [Google Scholar]
- Premature grey hair and hair loss among smokers: A new opportunity for health education? BMJ. 1996;313:1616.
- [CrossRef] [PubMed] [Google Scholar]
- Association of androgenetic alopecia with mortality from diabetes mellitus and heart disease. JAMA Dermatol. 2013;149:601-6.
- [CrossRef] [PubMed] [Google Scholar]
- Association between androgenetic alopecia and coronary artery disease in young male patients. Int J Trichology. 2014;6:5-7.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Definition of metabolic syndrome: Report of the national heart, lung, and blood institute/American Heart Association conference on scientific issues related to definition. Circulation. 2004;109:433-8.
- [CrossRef] [PubMed] [Google Scholar]
- Metabolic syndrome: Risk factor distribution and 18-year mortality in the multiple risk factor intervention trial. Diabetes Care. 2006;29:123-30.
- [CrossRef] [PubMed] [Google Scholar]
- Androgenetic alopecia as an indicator of metabolic syndrome and cardiovascular risk. Blood Press. 2016;25:141-8.
- [CrossRef] [PubMed] [Google Scholar]
- Pitx genes in development and disease. Cell Mol Life Sci. 2021;78:4921-38.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- The role of Pitx2 in maintaining the phenotype of myogenic precursor cells in the extraocular muscles. PLoS One. 2013;8:e58405.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Multiple roles of Pitx2 in cardiac development and disease. J Cardiovasc Dev Dis 2017:4.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- In silico assessment of genetic variation in PITX2 reveals the molecular mechanisms of calcium-mediated cellular triggered activity in atrial fibrillation. Annu Int Conf IEEE Eng Med Biol Soc 2020:2353-6.
- [CrossRef] [PubMed] [Google Scholar]
- Decoding the PITX2-controlled genetic network in atrial fibrillation. JCI Insight 2022:7.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Alopecia areata is associated with increased expression of heart disease biomarker cardiac troponin I. Acta Derm Venereol. 2018;98:776-82.
- [CrossRef] [PubMed] [Google Scholar]
- Female pattern hair loss and androgen excess: A report from the multidisciplinary androgen excess and PCOS committee. J Clin Endocrinol Metab. 2019;104:2875-91.
- [CrossRef] [PubMed] [Google Scholar]
- Androgenetic alopecia: An update. Indian J Dermatol Venereol Leprol. 2013;79:613-25.
- [CrossRef] [PubMed] [Google Scholar]
- The role of Pitx2 during cardiac development. Linking left-right signaling and congenital heart diseases. Trends Cardiovasc Med. 2003;13:157-63.
- [CrossRef] [PubMed] [Google Scholar]
- Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat Genet. 1996;14:392-9.
- [CrossRef] [PubMed] [Google Scholar]
- Identification of a Wnt/Dvl/beta-Catenin --> Pitx2 pathway mediating cell-type-specific proliferation during development. Cell. 2002;111:673-85.
- [CrossRef] [PubMed] [Google Scholar]
- Beta-catenin directly regulates Islet1 expression in cardiovascular progenitors and is required for multiple aspects of cardiogenesis. Proc Natl Acad Sci U S A. 2007;104:9313-8.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Pitx2, a beta-catenin-regulated transcription factor, regulates the differentiation of outer root sheath cells cultured in vitro. J Dermatol Sci. 2009;54:6-11.
- [CrossRef] [PubMed] [Google Scholar]






