Translate this page into:
Association of protein tyrosine phosphatase, non-receptor type 22 +1858C→T polymorphism and susceptibility to vitiligo: Systematic review and meta-analysis
Correspondence Address:
Harish Changotra
Department of Biotechnology and Bioinformatics, Jaypee University of Information Technology, Waknaghat, Solan - 173 234, Himachal Pradesh
India
| How to cite this article: Agarwal S, Changotra H. Association of protein tyrosine phosphatase, non-receptor type 22 +1858C→T polymorphism and susceptibility to vitiligo: Systematic review and meta-analysis. Indian J Dermatol Venereol Leprol 2017;83:183-189 |
Abstract
Background: Protein tyrosine phosphatase, non-receptor type 22 gene, which translates to lymphoid tyrosine phosphatase, is considered to be a susceptibility gene marker associated with several autoimmune diseases. Several studies have demonstrated the association of protein tyrosine phosphatase, non-receptor type 22 +1858C→T polymorphism with vitiligo. However, these studies showed conflicting results. Meta-analysis of the same was conducted earlier that included fewer number of publications in their study.Aim: We performed a meta-analysis of a total of seven studies consisting of 2094 cases and 3613 controls to evaluate the possible association of protein tyrosine phosphatase, non-receptor type 22 +1858C>T polymorphism with vitiligo susceptibility.
Methods: We conducted a literature search in PubMed, Google Scholar and Dogpile for all published paper on protein tyrosine phosphatase, non-receptor type 22 +1858C→T polymorphism and vitiligo risk till June 2016. Data analysis was performed by RevMan 5.3 and comprehensive meta-analysis v3.0 software.
Results: Meta-analysis showed an overall significant association of protein tyrosine phosphatase, non- receptor type 22 +1858C→T polymorphism with vitiligo in all models (allelic model [T vs. C]: odds ratio = 1.50, 95% confidence interval [1.32–1.71], P< 0.001; dominant model [TT + CT vs. CC]: odds ratio = 1.61, 95% confidence interval [1.16–2.24], P = 0.004; recessive model [TT vs. CT + CC]: odds ratio = 4.82, 95% confidence interval [1.11–20.92], P = 0.04; homozygous model [TT vs. CC]: odds ratio = 5.34, 95% confidence interval [1.23–23.24], P = 0.03; co-dominant model [CT vs. CC]: odds ratio = 1.52, 95% confidence interval [1.09–2.13], P = 0.01). No publication bias was detected in the funnel plot study.
Limitations: Limited ethnic-based studies, unable to satisfy data by gender or vitiligo-type are some limitations of the present meta-analysis.
Conclusion: Stratifying data by ethnicity showed an association of protein tyrosine phosphatase, non-receptor type 22 +1858C→T with vitiligo in European population (odds ratio = 1.53, 95% confidence interval [1.34–1.75], P< 0.001) but not in Asian population (odds ratio = 0.59, 95% confidence interval [0.26–1.32], P = 0.2). In conclusion, protein tyrosine phosphatase, non-receptor type 22 +1858 T allele predisposes European individuals to vitiligo.
Introduction
Vitiligo is a hypomelanotic dermatologic disorder on the skin that may gradually enlarge and appear anywhere on the body.[1],[2] It is caused by the destruction of functional melanocytes resulting in melanin loss from the lesion site. The etiology of melanocyte destruction has not been fully understood, but intricate interactions of environmental triggers, genetics, alteration in biochemical factors and immunological factors are thought to be involved in its pathogenesis.[3] Various hypotheses have been proposed, of which autoimmunity has been suggested to be playing a pivotal role in its pathogenesis. Epidemiological studies have reported the presence of autoantibodies against melanosomal proteins such as tyrosinase,[4] gp100/pmel17,[5] tyrosinase-related protein (tyrosinase-related protein 1 and tyrosinase-related protein 2)[6] in the serum and autoreactive cytotoxic T-cell lymphocytes in peripheral blood and perilesional skin of vitiligo patients,[7] which supports the autoimmunity theory of vitiligo. The autoimmune hypothesis is also supported because of the association of vitiligo with other autoimmune diseases such as rheumatoid arthritis, type 1 diabetes mellitus, pernicious anemia, autoimmune thyroid disease, Addison's disease and systemic lupus erythematosus.[8],[9],[10] Interaction between genetic and environmental factors influences the development of autoimmune disease. Linkage and association studies have been conducted through which many genes such as major histocompatibility complex, cytotoxic T-lymphocyte antigen 4, human leukocyte antigen, NALP1, protein tyrosine phosphatase, non-receptor type 22 and autoimmune regulator I, liver X receptor-α gene have been implicated in the pathogenesis of vitiligo.[1],[11],[12] Among these, protein tyrosine phosphatase, non-receptor type 22 (lymphoid-specific) locus is considered to be one of the strongest risk factors associated with autoimmune disease.[13] Protein tyrosine phosphatase, non-receptor type 22 gene located on chromosome 1p13.2-p13.1 codes for an intracellular protein named lymphoid-specific tyrosine phosphatase. Lymphoid-specific tyrosine phosphatase protein plays a crucial role in the negative regulation of T-cell antigen receptor signaling pathway.[14] Lymphoid-specific tyrosine phosphatase binds with the Src homology 3 domain of the C-terminal Src kinase forming a complex which inhibits the T cell receptor-signaling and ultimately inhibits T cell development and activation.[15] Protein tyrosine phosphatase, non-receptor type 22 gene maintains the central and peripheral tolerance in the body. Polymorphic change in protein tyrosine phosphatase, non-receptor type 22 at nucleotide position +1858 from C to T has shown to disrupt the function of protein tyrosine phosphatase, non-receptor type 22 which may predispose a person to autoimmune disease. Protein tyrosine phosphatase, non-receptor type 22 +1858C→T single-nucleotide polymorphism is a missense/nonsynonymous single-nucleotide polymorphism, in which change in the amino acid at position 620 from an arginine to tryptophan is observed (R620W). This change disrupts the interaction between lymphoid tyrosine phosphate and C-terminal Src kinase which downregulates the T-cell antigen receptor-signaling threshold. Thus, T-cell activation occurs in an uncontrollable fashion and may become a cause in the development of autoimmune disease.[13],[15] Protein tyrosine phosphatase, nonreceptor type 22 +1858C→T single-nucleotide polymorphism, first introduced by Mustelin's group,[16] has been associated with several autoimmune diseases such as type 1 diabetes mellitus,[16] Graves' disease,[17] systemic lupus erythematosus,[18] rheumatoid arthritis,[19] myasthenia gravis and [20] Addison's disease.[21]
Several studies were conducted to explore the association of protein tyrosine phosphatase, non-receptor type 22 +1858C→T and vitiligo susceptibility, but it still remains unclear because of inconsistent and inconclusive results from different studies.[8],[22],[23],[24],[25],[26],[27] This inconsistency may be due to inadequate statistical power, racial and ethnic differences, small sample size or other limitations. Meta-analysis of pooled data from different studies has been proven useful to assess an overall risk of a polymorphism with the disease susceptibility. Hence, we performed a meta-analysis of all available studies to clarify and to provide comprehensive assessment of the association between protein tyrosine phosphatase, non-receptor type 22 +1858C→T (rs2476601) and vitiligo susceptibility.
Methods
Strategy for literature search
To identify all articles that examined the association of protein tyrosine phosphatase, non-receptor type 22 +1858C→T polymorphism with vitiligo susceptibility, we conducted a literature search in PubMed, Google Scholar and Dogpile for all the published paper assessing the association between protein tyrosine phosphatase, non-receptor type 22 +1858C→T polymorphism and vitiligo risk using the following terms and keywords: “Protein tyrosine phosphatase, non-receptor type 22,” “protein tyrosine phosphatase, non-receptor type 22 1858C→T,” “rs2476601” and vitiligo. The last search updated to June 2016. Additional eligible studies were identified by a manual search of references cited in selected studies and previously published review articles on this topic.
Inclusion and exclusion criteria
Studies were considered to be eligible only when they meet the following inclusion criteria: (1) Study must be evaluating the association between protein tyrosine phosphatase, non-receptor type 22 +1858C→T polymorphism and vitiligo risk, (2) only case–control design studies were selected without gender bias, (3) study must have sufficient published data available such as sample size, genotype distribution or allelic frequencies for both cases and control for estimating the odds ratio with its 95% confidence interval, (4) genotype distribution of control group must be consistent with Hardy–Weinberg equilibrium and (5) family-based studies were excluded.
Data extraction
From each eligible study following data were extracted:First author's name, publication year, country of origin, ethnicity and genotyping distribution for the gene protein tyrosine phosphatase, nonreceptor type 22 in cases and control and P value of Hardy–Weinberg equilibrium. The genotype distribution for the gene polymorphism protein tyrosine phosphatase, nonreceptor type 22 +1858C→T in cases and control for each study is extracted for statistical analysis.
Statistical analysis
The strength of the association between protein tyrosine phosphatase, nonreceptor type 22 +1858C→T polymorphism and vitiligo risk was estimated by calculating pooled odds ratio and 95% confidence interval whose significance was determined by Z-test (P < 0.05 was considered statistically significant). The pooled odds ratios and 95% confidence intervals were calculated for the following genotypic models: Allelic model (T vs. C), homozygote model (TT vs. CC), dominant model (CT + TT vs. CC), recessive model (TT vs. CT + CC) and codominant model (CT vs. CC).
Heterogeneity test (Chi-square-based Q-test and I2-test) was performed to test the heterogeneity between studies which helps in selecting the appropriate model (random effect model or fixed effect model) to calculate the pooled effect estimate.[28] The Q-statistic tests with P < 0.10 or I2 >50% defined as the presence of a significant degree of heterogeneity between studies. For P > 0.10 or I2 <50%, fixed-effect model [29] was used; otherwise, random-effect model [30] was used to calculate the pooled effect. Sensitivity analysis was tested to study the effect of each study on the overall meta-analysis result and thus to conclude about the stability of meta-analysis results. The potential publication bias was estimated by visual inspection of the funnel plot along with the P value obtained by Begg's test and Egger's linear regression test. P < 0.05 was considered as the existence of statistically significant publication bias. All statistical tests were performed with software RevMan v5.3 (The Nordic Cochrane Centre, Copenhagen) and comprehensive meta-analysis v3.0.
Results
To assess the possible association of protein tyrosine phosphatase, nonreceptor type 22 +1858C>T polymorphism with vitiligo susceptibility, meta-analysis was done by combining all the available studies which fulfilled the inclusion criteria.
Search results and study characteristics
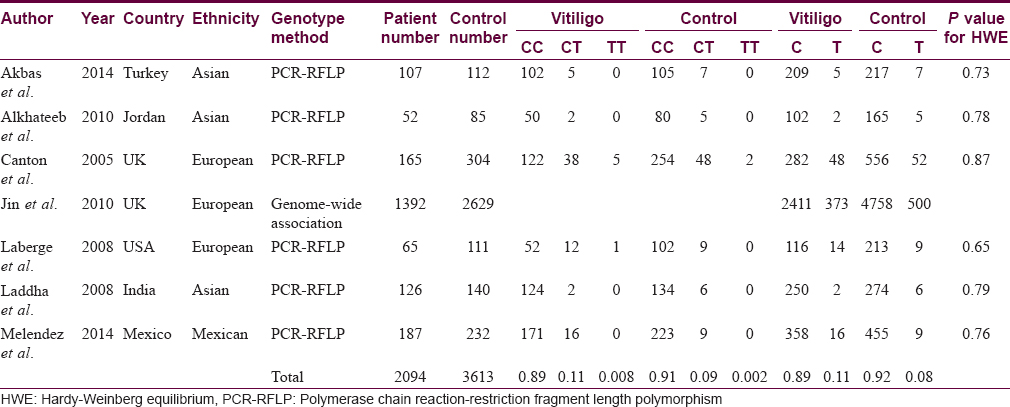
We identified 162 articles, out of which seven studies with total 2094 vitiligo patients and 3613 controls fulfilled the inclusion criteria and were included in this meta-analysis [Figure - 1]. Baseline characteristics and genotype distribution of included studies are shown in [Table - 1]. The genotype distributions in controls in each study were consistent with Hardy–Weinberg Equilibrium.
 |
| Figure 1: Flow chart of literature search, inclusion and exclusion criteria for meta-analysis |
Meta-Analysis Results
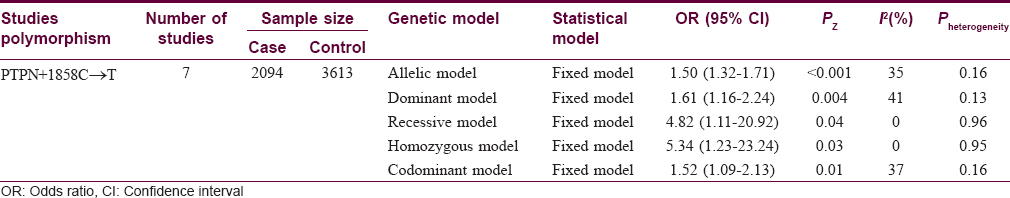
The pooled statistical results of meta-analysis of all the seven included studies to estimate the association of protein tyrosine phosphatase, nonreceptor type 22 +1858C→T with vitiligo risk are shown in [Table - 2]. No significant heterogeneity between the studies was found (P > 0.1, I2 < 50%); therefore, fixed effect model was used to calculate the pooled estimate. We found an overall significant association between the protein tyrosine phosphatase, nonreceptor type 22 +1858C→T and vitiligo risk under: Allelic model (1.50, 95% confidence interval [1.32–1.71] Z = 6.61, P < 0.001), dominant model (1.61, 95% confidence interval [1.16–2.24] Z = 2.84, P = 0.004), recessive model (4.82, 95% confidence interval [1.11–20.92] Z = 2.10, P = 0.04), homozygote model (5.34, 95% confidence interval [1.23–23.24] Z = 2.23, P = 0.03) while there was no significant association for codominant model (1.52, 95% confidence interval [1.09–2.13] Z = 2.45, P = 0.01) [Figure 2a], [Figure 2b], [Figure 2c], [Figure 2d], [Figure 2e]. In subgroup analysis stratified by ethnicity, we found a significant association between protein tyrosine phosphatase, nonreceptor type 22 +1858C→T and vitiligo risk in European population (1.53, 95% confidence interval [1.34–1.75] Z = 6.24, P < 0.001) but not with Asian (0.59, 95% confidence interval [0.26–1.32] Z = 1.28, P = 0.2) [Figure 3a] and [Figure 3b].
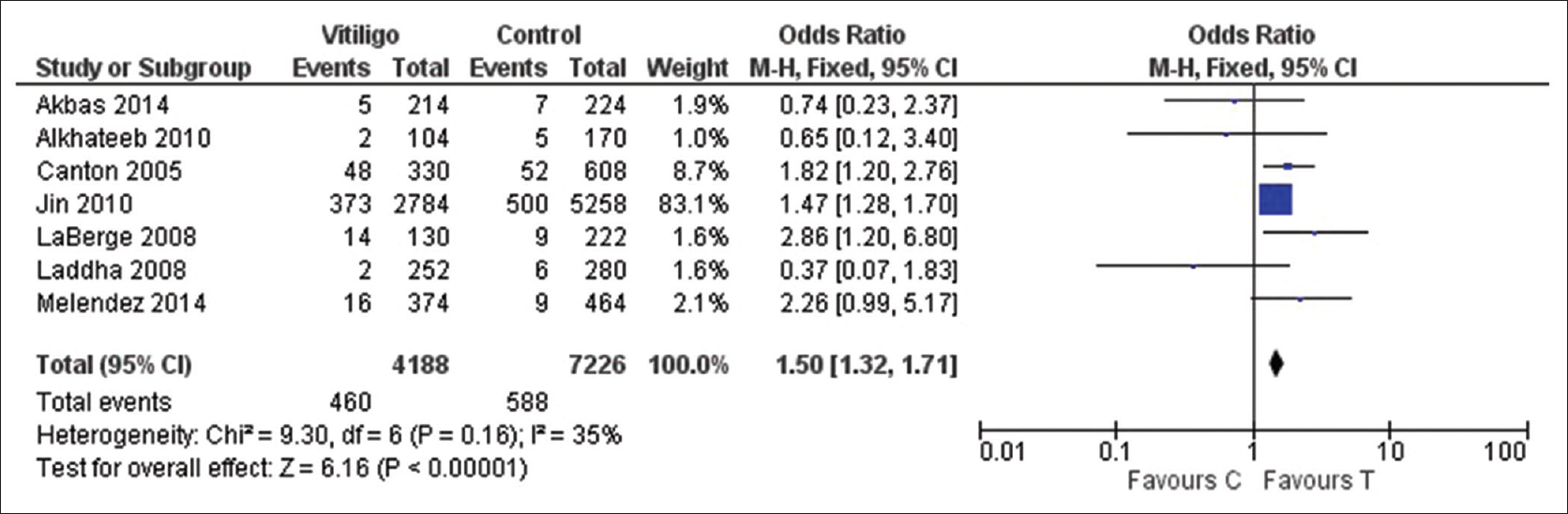 |
| Figure 2a: Forest plots to assess the association between the protein tyrosine phosphatase, nonreceptor type 22 +1858C→T and vitiligo risk collectively in all the studies included in the meta-analysis. The squares and horizontal lines correspond to odds ratio and 95% confidence interval of specific study and the area of squares reflects study weightage (inverse of the variance). The diamond represents the pooled odds ratio and its 95% confidence interval. Meta-analysis under allelic model, showed association of protein tyrosine phosphatase, nonreceptor type 22 +1858C→T with vitiligo risk |
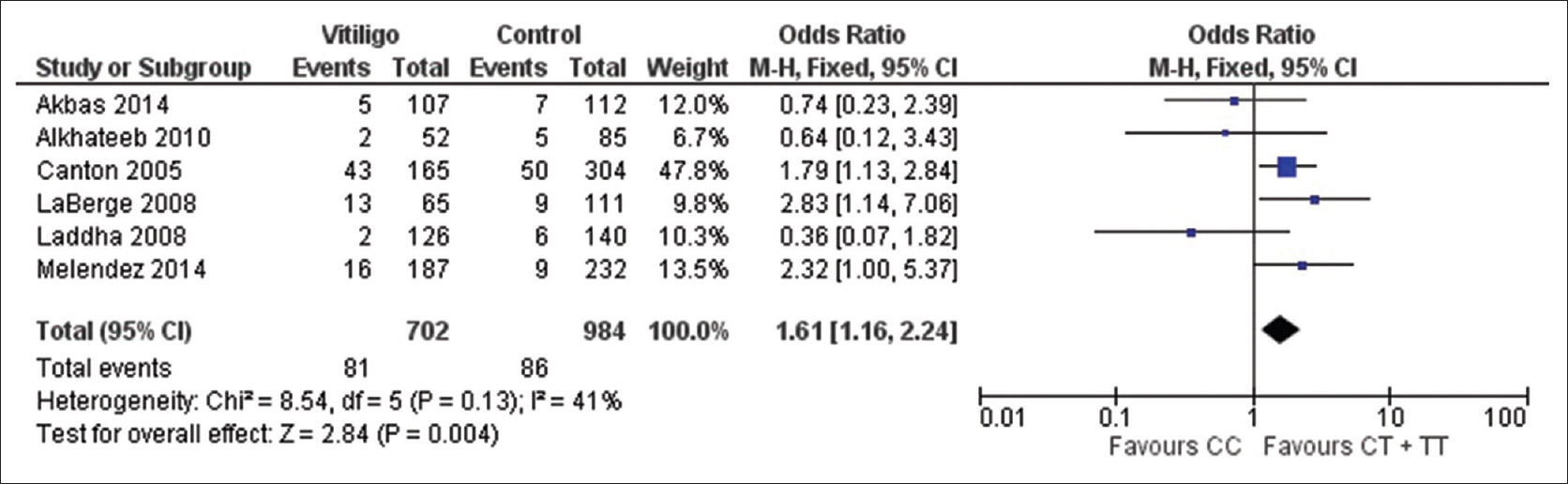 |
| Figure 2b: Forest plots to assess the association between the protein tyrosine phosphatase, nonreceptor type 22 +1858C→T and vitiligo risk collectively in all the studies included in the meta-analysis. The squares and horizontal lines correspond to odds ratio and 95% confidence interval of specific study and the area of squares reflects study weightage (inverse of the variance). The diamond represents the pooled odds ratio and its 95% confidence interval. Meta-analysis under dominant model, showed association of protein tyrosine phosphatase, nonreceptor type 22 +1858C→T with vitiligo risk |
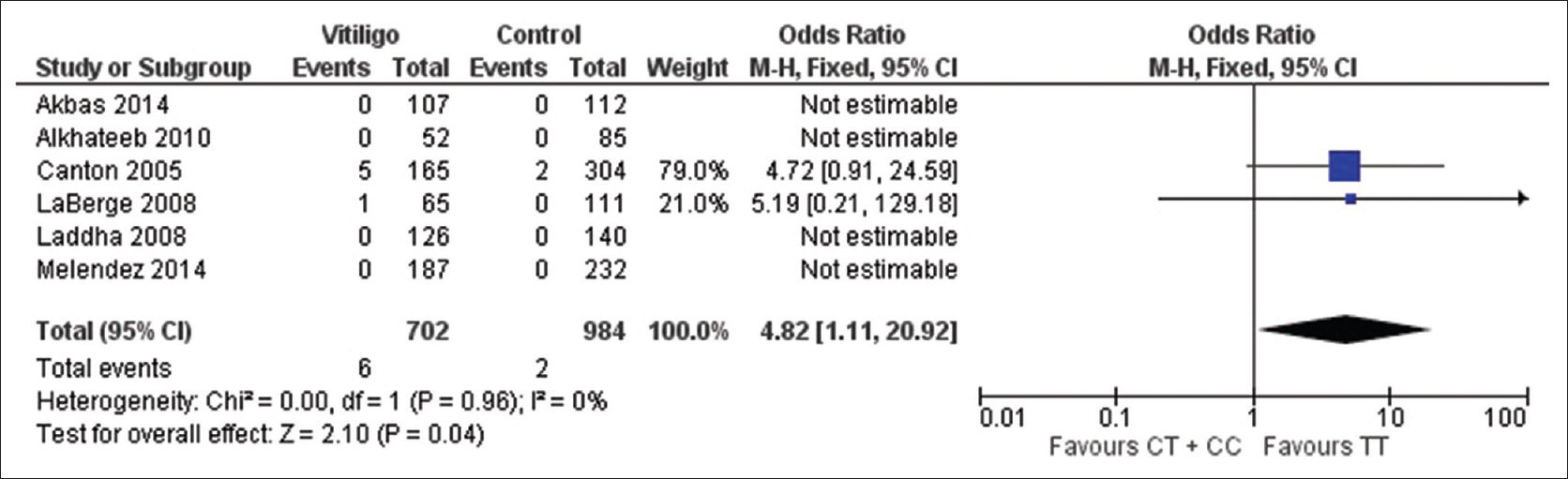 |
| Figure 2c: Forest plots to assess the association between the protein tyrosine phosphatase, nonreceptor type 22 +1858C→T and vitiligo risk collectively in all the studies included in the meta-analysis. The squares and horizontal lines correspond to odds ratio and 95% confidence interval of specific study and the area of squares reflects study weightage (inverse of the variance). The diamond represents the pooled odds ratio and its 95% confidence interval. Meta-analysis under recessive model, showed association of protein tyrosine phosphatase, nonreceptor type 22 +1858C→T with vitiligo risk |
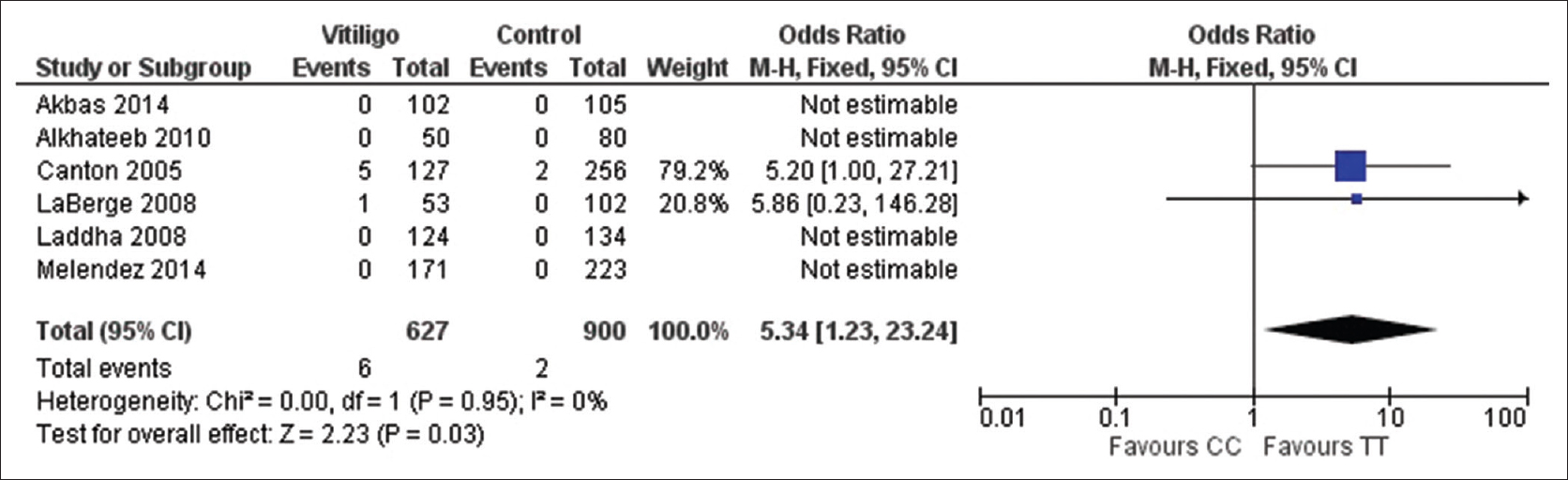 |
| Figure 2d: Forest plots to assess the association between the protein tyrosine phosphatase, nonreceptor type 22 +1858C.T and vitiligo risk collectively in all the studies included in the meta-analysis. The squares and horizontal lines correspond to odds ratio and 95% confidence interval of specific study and the area of squares reflects study weightage (inverse of the variance). The diamond represents the pooled odds ratio and its 95% confidence interval. Meta-analysis under homozygous model, showed association of protein tyrosine phosphatase, nonreceptor type 22 +1858C.T with vitiligo risk |
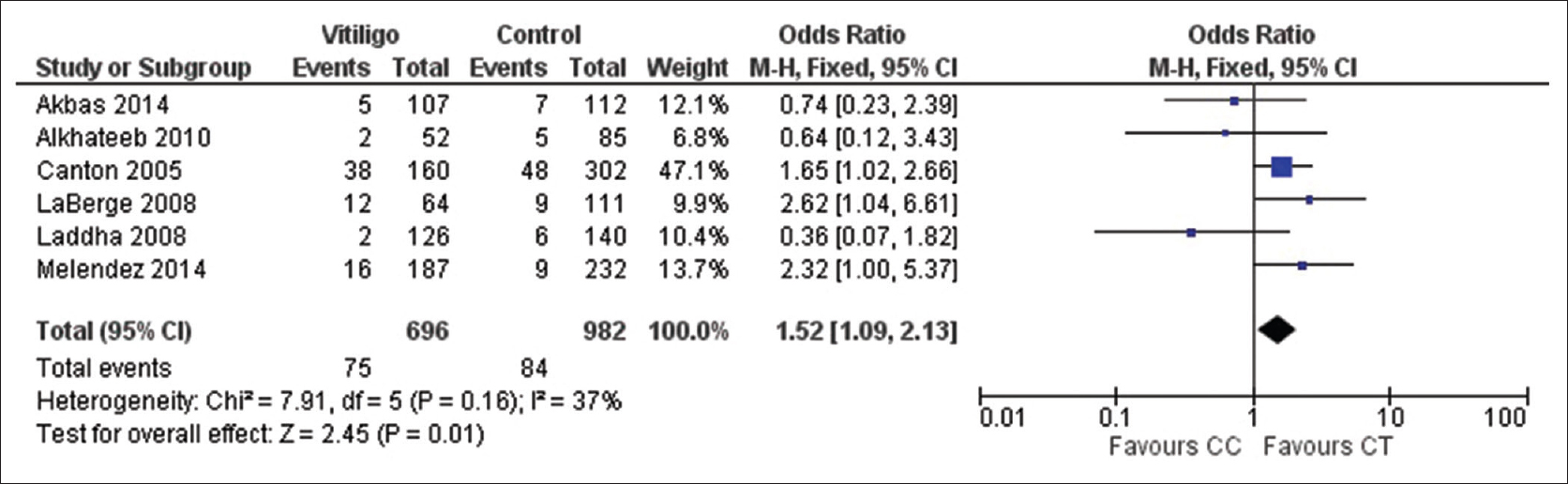 |
| Figure 2e: Forest plots to assess the association between the protein tyrosine phosphatase, nonreceptor type 22 +1858C→T and vitiligo risk collectively in all the studies included in the meta-analysis. The squares and horizontal lines correspond to odds ratio and 95% confidence interval of specific study and the area of squares reflects study weightage (inverse of the variance). The diamond represents the pooled odds ratio and its 95% confidence interval. Meta-analysis under co-dominant model, showed association of protein tyrosine phosphatase, nonreceptor type 22 +1858C→T with vitiligo risk |
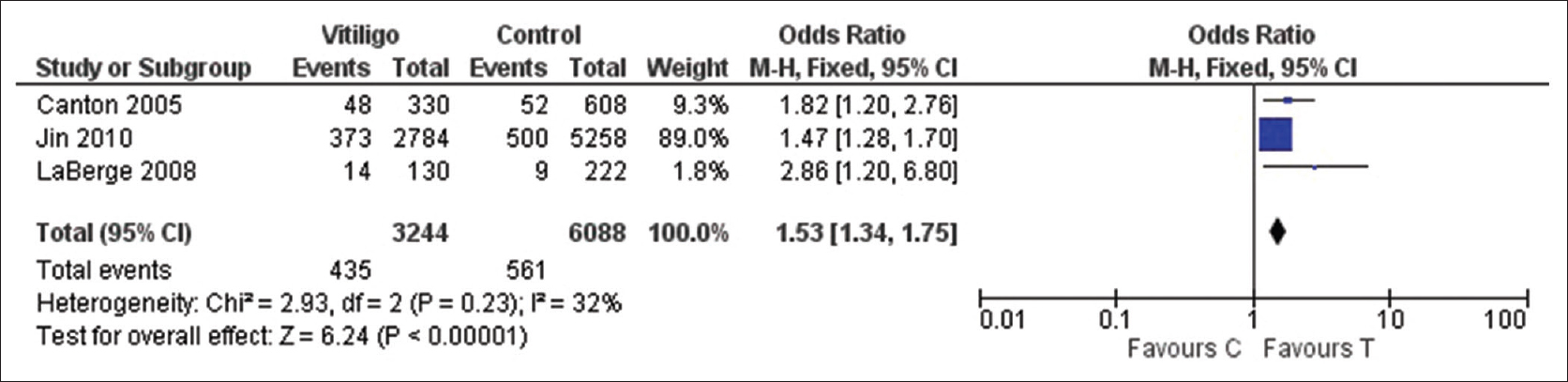 |
| Figure 3a: Forest plots to assess the association between the protein tyrosine phosphatase, nonreceptor type 22 +1858C→T and vitiligo risk according to the ethnicity, i.e., European, under allelic model |
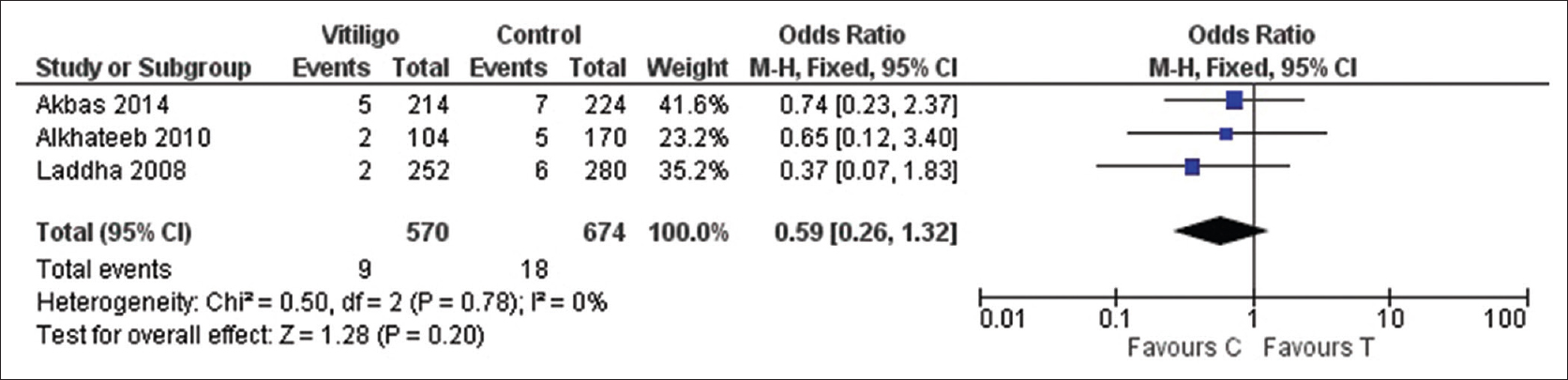 |
| Figure 3b: Forest plots to assess the association between the protein tyrosine phosphatase, nonreceptor type 22 +1858C→T and vitiligo risk according to the ethnicity, i.e., Asian, under allelic model |
Sensitivity analysis and publication bias
Odds ratio and corresponding 95% confidence interval were not significantly altered after removing each study which confirms the stability of meta-analysis results. There was a lack of publication bias as shown by symmetry of inverted funnel plot [Figure - 4]. To confirm publication bias, both Begg's test and Egger's regression test were performed. P values for both tests support no existence of publication bias under the models: Allelic (Begg's P = 0.45, Egger's P = 0.73); dominant (Begg's P = 0.35, Egger's P = 0.17); recessive (not estimable); codominant ( Begg's P = 0.35, Egger's P = 0.19); homozygous (not estimable).
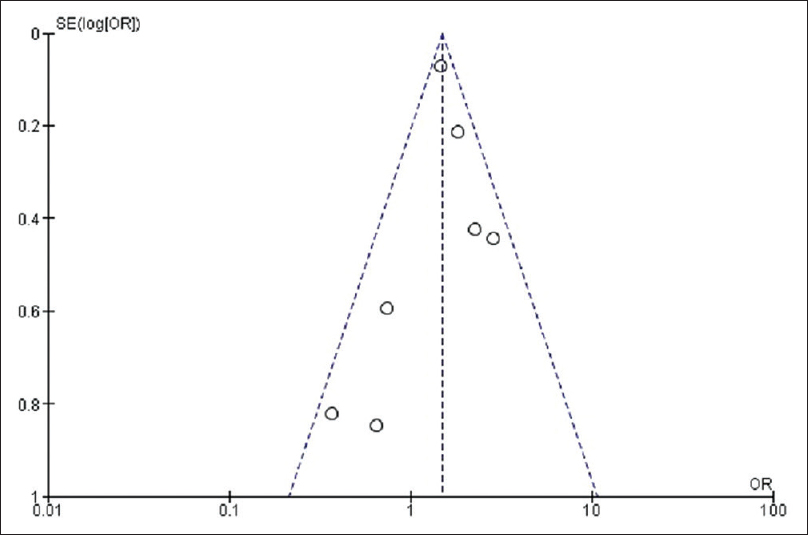 |
| Figure 4: Funnel plot to assess the publication bias. Odds ratio is plotted against the standard error of log odds ratio for studies on protein tyrosine phosphatase, nonreceptor type 22 +1858C→T polymorphism |
Discussion
Protein tyrosine phosphatase, nonreceptor type 22 gene encoding for lymphoid tyrosine phosphatase protein, forms a complex with C-terminal Src kinase protein, is known to play a key role in the regulation of T-cell activation and development by inhibiting the T-cell receptor-signaling pathway and thus involved in the maintenance of central and peripheral tolerance in the body.[14] Protein tyrosine phosphatase, nonreceptor type 22 gene knockout experiments have demonstrated the elevated titers of T-cell-dependent antibodies, immunoglobulin G1 and immunoglobulin G2.[7] Variation in gene sequence such as +1858C→T variant has been found to be associated with overactivity of T-cell and may predispose an individual to autoimmune disease. Previous studies along with meta-analysis reports have shown that protein tyrosine phosphatase, nonreceptor type 22 +1858C→T has been associated with the number of autoimmune diseases.[5],[17],[18],[31],[32] The autoimmune nature of vitiligo has been supported by the presence of circulating melanocyte antibodies in human and animal models and its association with other autoimmune diseases. Based on this information, protein tyrosine phosphatase, nonreceptor type 22 polymorphism has been thought to be associated with vitiligo pathogenesis. Several studies in the different ethnic background were conducted to assess the association of protein tyrosine phosphatase, nonreceptor type 22 +1858C→T with vitiligo susceptibility,[8],[22],[23],[24],[25],[26],[27] but inconsistent and inconclusive results were obtained. Studies by Cantón et al., LaBerge et al. and Jin et al. reported association between the protein tyrosine phosphatase, nonreceptor type 22 +1858C→T polymorphism with vitiligo susceptibility among English, Romanian and English-North American population, respectively.[23],[25],[26] However, Laddha et al., Alkhateeb et al. and Akbas et al. reported no association between the protein tyrosine phosphatase, nonreceptor type 22 +1858C→T polymorphism with vitiligo susceptibility among Indian, Jordanian and Turkish population, respectively.[22],[27],[33] Garcia-Melendez et al. reported the association of protein tyrosine phosphatase, nonreceptor type 22 T allele with active vitiligo individual in the Mexican population.[24] Alkhateeb et al. suggested the association of the mutant variant with active vitiligo, but because of the limited number of patients, could not conclude their results.[8]
A total of nine studies were identified by systematic review of literature search, but two of the studies did not fulfill the inclusion criteria as mentioned in [Figure - 1]. Meta-analysis of seven studies fulfilling inclusion criteria was performed to evaluate the association of protein tyrosine phosphatase, nonreceptor type 22 +1858C→T with vitiligo susceptibility with the aim of obtaining single conclusive result.[8],[22],[23],[24],[25],[26],[27] Our meta-analysis showed an overall significant association of protein tyrosine phosphatase, nonreceptor type 22 +1858C→T with vitiligo susceptibility under all genetic and allelic models suggesting that the protein tyrosine phosphatase, nonreceptor type 22 T allele is the risk factor in vitiligo pathogenesis. Results of this meta-analysis are inconsistent with the previous meta-analysis result conducted by Song et al.[34] In their study, only five available studies were included with the data obtained only from two ethnic populations, i.e., Asian and European. Two new case–control studies have been published after this meta-analysis.[22],[24] Hence, to provide a comprehensive assessment of the association; we performed an updated meta-analysis of all available studies. Addition of these studies in our meta-analysis provided the advantage of (1) addition of new ethnic population, i.e., American in the meta-analysis which was lacking in the Song et al.,[34] (2) increase in the sample size resulting in more powerful statistical evidence and (3) reduction in the heterogeneity. Frequency of the risk allele (+1858T) varies in different populations and these associations could be related to these frequencies. Therefore, on stratifying data by ethnicity revealed that the protein tyrosine phosphatase, nonreceptor type 22 +1858C→T polymorphism is associated with vitiligo susceptibility in European population only and not in Asian and American populations. The drawbacks of the present meta-analysis are (1) limited number of ethnic-based studies for protein tyrosine phosphatase, nonreceptor type 22 polymorphism, (2) small sample size and (3) data were not stratified by gender or vitiligo type because of insufficient information.
Conclusion
In conclusion, meta-analysis results showed that the protein tyrosine phosphatase, nonreceptor type 22 +1858T-allele predisposes an European individual to vitiligo and thus uncovers the genetic factors involved in the pathogenesis of vitiligo. We suggest that greater number of ethnicity-based studies with more number of samples is required to further confirm this association.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
| 1. |
Shajil EM, Chatterjee S, Agrawal D, Bagchi T, Begum R. Vitiligo: Pathomechanisms and genetic polymorphism of susceptible genes. Indian J Exp Biol 2006;44:526-39.
[Google Scholar]
|
| 2. |
Taïeb A, Picardo M. Clinical practice. Vitiligo. N Engl J Med 2009;360:160-9.
[Google Scholar]
|
| 3. |
Gopal KV, Rama Rao GR, Kumar YH, Appa Rao MV, Vasudev P; Srikant. Vitiligo: A part of a systemic autoimmune process. Indian J Dermatol Venereol Leprol 2007;73:162-5.
[Google Scholar]
|
| 4. |
Kemp EH, Emhemad S, Akhtar S, Watson PF, Gawkrodger DJ, Weetman AP. Autoantibodies against tyrosine hydroxylase in patients with non-segmental (generalised) vitiligo. Exp Dermatol 2011;20:35-40.
[Google Scholar]
|
| 5. |
Kemp EH, Gawkrodger DJ, Watson PF, Weetman AP. Autoantibodies to human melanocyte-specific protein pmel17 in the sera of vitiligo patients: A sensitive and quantitative radioimmunoassay (RIA). Clin Exp Immunol 1998;114:333-8.
[Google Scholar]
|
| 6. |
Kemp EH, Waterman EA, Gawkrodger DJ, Watson PF, Weetman AP. Autoantibodies to tyrosinase-related protein-1 detected in the sera of vitiligo patients using a quantitative radiobinding assay. Br J Dermatol 1998;139:798-805.
[Google Scholar]
|
| 7. |
Middelkamp-Hup MA, Bos JD, Rius-Diaz F, Gonzalez S, Westerhof W. Treatment of vitiligo vulgaris with narrow-band UVB and oral Polypodium leucotomos extract: A randomized double-blind placebo-controlled study. J Eur Acad Dermatol Venereol 2007;21:942-50.
[Google Scholar]
|
| 8. |
Alkhateeb A, Qarqaz F, Al-Sabah J, Al Rashaideh T. Clinical characteristics and PTPN22 1858C/T variant analysis in Jordanian Arab vitiligo patients. Mol Diagn Ther 2010;14:179-84.
[Google Scholar]
|
| 9. |
Laberge G, Mailloux CM, Gowan K, Holland P, Bennett DC, Fain PR, et al. Early disease onset and increased risk of other autoimmune diseases in familial generalized vitiligo. Pigment Cell Res 2005;18:300-5.
[Google Scholar]
|
| 10. |
Tarlé RG, Nascimento LM, Mira MT, Castro CC. Vitiligo – Part 1. An Bras Dermatol 2014;89:461-70.
[Google Scholar]
|
| 11. |
Spritz RA. The genetics of generalized vitiligo and associated autoimmune diseases. Pigment Cell Res 2007;20:271-8.
[Google Scholar]
|
| 12. |
Agarwal S, Kaur G, Randhawa R, Mahajan V, Bansal R, Changotra H. Liver X Receptor-a polymorphisms (rs11039155 and rs2279238) are associated with susceptibility to vitiligo. Meta Gene 2016;8:33-6.
[Google Scholar]
|
| 13. |
Burn GL, Svensson L, Sanchez-Blanco C, Saini M, Cope AP. Why is PTPN22 a good candidate susceptibility gene for autoimmune disease? FEBS Lett 2011;585:3689-98.
[Google Scholar]
|
| 14. |
Vang T, Miletic AV, Bottini N, Mustelin T. Protein tyrosine phosphatase PTPN22 in human autoimmunity. Autoimmunity 2007;40:453-61.
[Google Scholar]
|
| 15. |
Cloutier JF, Veillette A. Cooperative inhibition of T-cell antigen receptor signaling by a complex between a kinase and a phosphatase. J Exp Med 1999;189:111-21.
[Google Scholar]
|
| 16. |
Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, Rostamkhani M, et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet 2004;36:337-8.
[Google Scholar]
|
| 17. |
Velaga MR, Wilson V, Jennings CE, Owen CJ, Herington S, Donaldson PT, et al. The codon 620 tryptophan allele of the lymphoid tyrosine phosphatase (LYP) gene is a major determinant of Graves' disease. J Clin Endocrinol Metab 2004;89:5862-5.
[Google Scholar]
|
| 18. |
Kyogoku C, Langefeld CD, Ortmann WA, Lee A, Selby S, Carlton VE, et al. Genetic association of the R620W polymorphism of protein tyrosine phosphatase PTPN22 with human SLE. Am J Hum Genet 2004;75:504-7.
[Google Scholar]
|
| 19. |
Begovich AB, Carlton VE, Honigberg LA, Schrodi SJ, Chokkalingam AP, Alexander HC, et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet 2004;75:330-7.
[Google Scholar]
|
| 20. |
Vandiedonck C, Capdevielle C, Giraud M, Krumeich S, Jais JP, Eymard B, et al. Association of the PTPN22*R620W polymorphism with autoimmune myasthenia gravis. Ann Neurol 2006;59:404-7.
[Google Scholar]
|
| 21. |
Skinningsrud B, Husebye ES, Gervin K, Løvås K, Blomhoff A, Wolff AB, et al. Mutation screening of PTPN22: Association of the 1858T-allele with Addison's disease. Eur J Hum Genet 2008;16:977-82.
[Google Scholar]
|
| 22. |
Akbas H, Dertlioglu SB, Dilmec F, Atay AE. Lack of association between PTPN22 Gene +1858 C→T polymorphism and susceptibility to generalized vitiligo in a Turkish population. Ann Dermatol 2014;26:88-91.
[Google Scholar]
|
| 23. |
Cantón I, Akhtar S, Gavalas NG, Gawkrodger DJ, Blomhoff A, Watson PF, et al. A single-nucleotide polymorphism in the gene encoding lymphoid protein tyrosine phosphatase (PTPN22) confers susceptibility to generalised vitiligo. Genes Immun 2005;6:584-7.
[Google Scholar]
|
| 24. |
Garcia-Melendez ME, Salinas-Santander M, Sanchez-Dominguez C, Gonzalez-Cardenas H, Cerda-Flores RM, Ocampo-Candiani J, et al. Protein tyrosine phosphatase PTPN22 +1858C/T polymorphism is associated with active vitiligo. Exp Ther Med 2014;8:1433-7.
[Google Scholar]
|
| 25. |
Jin Y, Birlea SA, Fain PR, Gowan K, Riccardi SL, Holland PJ, et al. Variant of TYR and autoimmunity susceptibility loci in generalized vitiligo. N Engl J Med 2010;362:1686-97.
[Google Scholar]
|
| 26. |
Laberge GS, Birlea SA, Fain PR, Spritz RA. The PTPN22-1858C→T (R620W) functional polymorphism is associated with generalized vitiligo in the Romanian population. Pigment Cell Melanoma Res 2008;21:206-8.
[Google Scholar]
|
| 27. |
Laddha NC, Dwivedi M, Shajil EM, Prajapati H, Marfatia YS, Begum R. Association of PTPN22 1858C/T polymorphism with vitiligo susceptibility in Gujarat population. J Dermatol Sci 2008;49:260-2.
[Google Scholar]
|
| 28. |
Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med 2002;21:1539-58.
[Google Scholar]
|
| 29. |
Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 1959;22:719-48.
[Google Scholar]
|
| 30. |
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials 1986;7:177-88.
[Google Scholar]
|
| 31. |
Dong F, Yang G, Pan HW, Huang WH, Jing LP, Liang WK, et al. The association of PTPN22 rs2476601 polymorphism and CTLA-4 rs231775 polymorphism with LADA risks: A systematic review and meta-analysis. Acta Diabetol 2014;51:691-703.
[Google Scholar]
|
| 32. |
Lee YH, Rho YH, Choi SJ, Ji JD, Song GG, Nath SK, et al. The PTPN22 C1858T functional polymorphism and autoimmune diseases – A meta-analysis. Rheumatology (Oxford) 2007;46:49-56.
[Google Scholar]
|
| 33. |
Alkhateeb A, Fain PR, Thody A, Bennett DC, Spritz RA. Epidemiology of vitiligo and associated autoimmune diseases in Caucasian probands and their families. Pigment Cell Res 2003;16:208-14.
[Google Scholar]
|
| 34. |
Song GG, Kim JH, Lee YH. The CTLA-4+49 A/G, CT60 A/G and PTPN22 1858 C/T polymorphisms and susceptibility to vitiligo: A meta-analysis. Mol Biol Rep 2013;40:2985-93.
[Google Scholar]
|
Fulltext Views
4,189
PDF downloads
2,019





