Development of a novel loop-mediated isothermal amplification assay for rapid detection of Mycobacterium leprae in clinical samples
Corresponding author: Dr. Poonam Salotra, Molecular Parasitology Lab, ICMR-National Institute of Pathology, Safdarjung Hospital Campus, New Delhi - 110 029, India. poonamsalotra@hotmail.com
-
Received: ,
Accepted: ,
How to cite this article: Joshi S, Sharma V, Ramesh V, Singh R, Salotra P. Development of a novel loop-mediated isothermal amplification assay for rapid detection of Mycobacterium leprae in clinical samples. Indian J Dermatol Venereol Leprol 2021;87:491-7.
Abstract
Background:
Sensitive and definitive diagnostic tests are required for timely treatment of leprosy and to control its transmission.
Aim:
In the present study, we report the development of loop-mediated isothermal amplification assay using six primers targeting the RLEP gene sequence uniquely present in Mycobacterium leprae.
Methods:
Tissue punch samples (n = 50) and slit aspirates (n = 50) from confirmed cases of leprosy (M. leprae positive by quantitative polymerase chain reaction), reporting at the Department of Dermatology, Safdarjung Hospital, New Delhi, were analyzed using newly developed closed tube loop-mediated isothermal amplification assay. The sensitivity and specificity; positive predictive value, negative predictive value and accuracy were calculated using MedCalc statistical software.
Results:
The loop-mediated isothermal amplification assay specifically amplified M. leprae genomic DNA with an analytical sensitivity of 100 fg. About 47 Out of the 50 quantitative polymerase chain reactions confirmed M. leprae positive tissue samples, 47 were positive by loop-mediated isothermal amplification assay (sensitivity 94%; 95% confidence interval 83.5%–98.8%) while only 31/50 were positive by histopathology (sensitivity 62%; 95% confidence interval 47.2%–75.4%) . Using slit aspirate samples of these 50 patients, 42 were positive by both quantitative polymerase chain reaction and loop-mediated isothermal amplification assay (sensitivity 84%; 95% confidence interval 70.9%–92.8%) while only 23/50 (sensitivity 46%; 95% confidence interval 31.8%–60.7%) were positive by microscopy.
Limitations:
In the present study, the leprosy patient cohort was not uniform, as it comprised a lower number of paucibacillary cases (22%) compared to multibacillary (78%) cases.
Conclusion:
Loop-mediated isothermal amplification assay established here provides a rapid and accurate diagnostic test for leprosy in terms of sensitivity and specificity. The assay is simple to perform in comparison with other molecular techniques (polymerase chain reaction/quantitative polymerase chain reaction) and has potential for field applicability.
Keywords
Diagnosis
histopathology/microscopy
leprosy
loop-mediated isothermal amplification
quantitative polymerase chain reaction
Introduction
Leprosy caused by Mycobacterium leprae, an acid-fast bacterium, has been known since the biblical times with reports of cases dating over 3000 years ago.1 The disease is endemic in tropical countries with new cases being mainly reported from India (58%), Brazil (16%) and Indonesia (9%).2 According to the World Health Organization, the global prevalence at the end of 2016 was 171,948 with a registered prevalence rate of 0.23/10,000 individuals.3 In India, the leprosy prevalence had decreased markedly since the introduction of multidrug therapy, from a prevalence rate of 57.8/10,000 in 1983 to 0.84/10,000 in 2006; however, the rate of new case detection has increased.4 According to the National Leprosy Elimination Program, Government of India (2018), about 88,000 people in India are suffering from leprosy (prevalence rate = 0.66/10000) with 135,485 new cases reported in 2017 (annual new case detection = 10.17/10000).
The spread of leprosy is usually caused by frequent close contact for a longer duration between an untreated patient and a person genetically susceptible to developing the disease. Patients often experience numbness, skin lesions, muscle weakness, enlarged nerves and joint pain. It has been observed that leprosy is often misdiagnosed with other similar skin diseases of the infiltrated plaque, papules or nodules including cutaneous leishmaniasis, macular post-kala-azar dermal leishmaniasis, cutaneous tuberculosis, sarcoidosis, lymphoma and syphilis.5 Accurate, specific and sensitive methodologies are needed to provide a definite diagnosis. Presently, microscopic observation of Ziehl-Neelsen stained slides6,7 and histopathological examination of cutaneous lesion biopsy8,9 are considered as “gold standard” for the diagnosis of leprosy but are less sensitive, time-consuming and dependent on technical expertise. The molecular methodologies, although not routinely used, are sensitive and specific; however, these need sophisticated and expensive equipment that may not be available in laboratories with limited resources. There have been numerous studies based on molecular techniques such as polymerase chain reaction or nested polymerase chain reaction to detect M. leprae in tissue samples10-13 and in slit aspirate samples10,14,15 with variable sensitivity and specificity. The highly conserved repetitive sequence RLEP has been the preferred target due to its high copy number (n = 37),10,14 although other targets such as rpoT, 16S rRNA and Sod A have also been exploited for detection of M. leprae by polymerase chain reaction/ quantitative polymerase chain reaction.16,17 Several cost-effective isothermal amplification-based techniques have emerged to substitute expensive molecular methods.18 Amongst these, loop-mediated isothermal amplification is simple, rapid, specific and sensitive and only a heating block or water bath capable to maintain a constant temperature (60 ̊C to 65 ºC) is required.19 The technique has many advantages such as the reaction proceeds isothermally,19 crude DNA extracts can be used directly without purification,19,20 and the products can be detected visually using multiple parameters including turbidity, fluorescence and color. For naked-eye detection, intercalating dye such as SYBR Green I, calcein or malachite green is added to the amplified products.21,22 The present study is focused to develop leprosy specific loop-mediated isothermal amplification assay targeting the RLEP gene to detect M. leprae in clinical samples.
Methods
Parasite DNA samples
To examine the specificity of the designed primers, M. leprae genomic DNA (NR-19350), obtained from BEI Resources, VA, USA, was tested along with extracted DNA from various co-endemic disease-causing bacteria/ parasites such as Mycobacterium tuberculosis, Acinetobacter baumannii, Klebsiella pneumoniae, Leishmania donovani, Leishmania major, Leishmania tropica, Plasmodium vivax and Plasmodium falciparum used as negative controls. Once the novel loop-mediated isothermal amplification assay for M. leprae detection was established, it was applied to the clinical samples of leprosy patients, while samples from other skin diseases were used as controls to determine the diagnostic specificity of the assay.
Clinical samples
The study was conducted after obtaining ethical clearance under the guidelines of the International Ethics Committee of Safdarjung Hospital, New Delhi and informed consent of the participants. At pretreatment stage, 3 mm punch tissue sample from skin lesions (n = 50) and slit aspirates taken from standard sites i.e., bilaterally at the eyebrows, earlobes and additionally selected site that observed skin lesions (n = 50) in 200μL NET buffer (150 mmol/L NaCl, 15 mmol/L Tris-HCl [pH 8.3] and 1 mmol/L EDTA) were collected from leprosy patients presenting characteristic clinical manifestations, reporting at the dermatology department of our hospital from June 2017 till June 2018. All cases were confirmed by quantitative polymerase chain reaction using tissue samples. Tissue samples from other skin diseases including post-kala-azar dermal leishmaniasis, vitiligo, sporotrichosis and pityriasis lichenoides chronica, lichen sclerosis, pityriasis rosea (n = 25) and normal skin samples (n = 15) were used as negative controls. Patients positive for HIV, hepatitis B and C, tuberculosis or any other systemic ailments were excluded. Pregnant or lactating women were excluded from the study. For DNA isolation, QIAamp DNA mini kit was used following manufacturer’s instructions. The isolated tissue DNA were eluted in 50μL (≈ 170 ng/μL) and slit aspirate DNA in 20μL (≈ 3.5 ng/μL) of nuclease-free water and stored at -30°C till further use.
Quantitative real-time polymerase chain reaction
The quantitative polymerase chain reaction was performed using primers described in an earlier study [Table 1 and Figure 1].11 Briefly, M. leprae specific real-time polymerase chain reaction was set up in an ABI Prism 7500 sequence detection system (Applied Biosystems, USA) using RLEP based forward and reverse primers. The quantitative polymerase chain reaction was performed as per the manufacturer’s instructions in a 10 μL reaction mixture consisting of 1X SYBR Green Fast polymerase chain reaction Master mix (Applied Biosystems, USA), 5 pmol forward primer, 5 pmol reverse primers and 1 μL volume of DNA from the sample. The results were obtained in accordance with the first fluorescent signal detection cycle threshold and the sample was considered positive when it showed cycle threshold smaller than 34 (negative cutoff value of >34). The cycling parameters included 50 °C for 2 min, 95 °C for 10 min and 40 cycles of 95 °C for 15 s and 61 °C for 60 s. The analytical sensitivity of SYBR Green I based RLEP specific quantitative polymerase chain reaction assay was determined using serially diluted (tenfold; 1 ng/μL to 1 fg/μL) M. leprae genomic DNA.
| Name of primer | Primer sequence (5’-3’) | Reference |
|---|---|---|
| qPCR primer | ||
| RLEP-forward | TGCATGTCATGGCCTTGAGG | 11 |
| RLEP-reverse | CACCGATACCAGCGGCAGAA | |
| LAMP primer | ||
| RLEP-F3 | TTGTTGGTGGGTGGCTGA | This study |
| RLEP-B3 | CGGCGCTAACAACTATCCTC | |
| RLEP-FIP (F1c-F2) | TTACGTGCGCCGCGCTAATCCTGCTTTCGATGAGGCTTCG | |
| RLEP-BIP (B1c-B2) | GGTGGATGCTGCTTGGTCTACATGCATCGATATCGCCTTCAG | |
| RLEP-FLP | CACTGCGGCAAAGCACA | |
| RLEP-BLP | TGTTGATGATGCCAGGGGC |
FIP: forward inner primer, BIP: backward inner primer, F3: outer forward primer, B3: outer backward primer, FLP: forward loop primer, BLP: backward loop primer, qPCR: quantitative polymerase chain reaction, LAMP: loop-mediated isothermal amplification

-
RLEP gene (Accession no X17153.1) sequence showing six primers (outer forward primer, outer backward primer, forward inner primer, backward inner primer, forward loop primer and backward loop primer) for loop-mediated isothermal amplification assay; Forward and reverse primers for quantitative polymerase chain reaction
Loop-mediated isothermal amplification assay
Loop-mediated isothermal amplification assay was developed targeting repetitive and specific RLEP gene (accession no X17153.1) sequence uniquely present in M. leprae. The six primers; forward inner primer, backward inner primer, outer forward primer, outer backward primer, forward loop primer and backward loop primer were designed using Primer Explorer V4 software (http://primerexplorer.jp/e/) [Table 1 and Figure 1]. Loop-mediated isothermal amplification reaction was performed as described earlier22 in 25μL reaction mixture containing 40 pmol each of forward inner primer and backward inner primer primers, 5 pmol each of outer forward primer and outer backward primer primers, 20 pmol each of the forward loop primer and backward loop primer, 1.4 mM of each deoxynucleoside triphosphate, 0.8 M betaine, 20 mM Tris-HCl, pH 8.8, 10 mM KCl, 10 mM (NH4)2SO4, 8 mM MgSO4, 0.1% TritonX-100, 8U Bst 2.0 (New England Biolabs, Massachusetts, USA) and 1–5 μL sample DNA at different time points (30 min to 120 min) at 65 °C in a heating block. For naked-eye detection, closed tube technology was used where SYBR Green I (Molecular Probes, Eugene, OR, USA) was added inside the tube cap at the beginning and a quick spin was given after completion of the assay to allow mixing of SYBR Green I with the amplified product. Green color in tube indicated positive while negative remained orange. Once the assay was established it was applied to clinical samples to determine diagnostic sensitivity of the assay. The assay was performed at 65 °C for varied time points ranging from 30 min to 120 min for different clinical samples. The reaction time for loop-mediated isothermal amplification assay was optimized as 60 min for tissue samples and 90 min for slit aspirate samples.
Statistical analysis
Samples were confirmed as M. leprae positive using quantitative polymerase chain reaction as a “gold standard” and classified as true positive, true negative, false positive and false negative for loop-mediated isothermal amplification assay evaluation. The clinical sensitivity, specificity, 95% confidence interval and accuracy were calculated for loop-mediated isothermal amplification using MedCalc statistical software. The positive predictive value was calculated as (number of true positives)/(number of true positives + number of false positives) ×100 and the negative predictive value was calculated as (number of true negatives)/(number of true negatives + number of false negatives) ×100. The accuracy was calculated as (number of true positives + number of true negatives)/(total number of patients) ×100.
Results
The average age of patients (n = 50) was 36 years (14–72 years) with men: women ratio is 2.8:1 (37 men and 13 women patients). Amongst the leprosy patients, on the basis of the number of lesions present all over the body, 39 patients were characterized as multibacillary presenting more than five lesions, while 11 as paucibacillary with less than five lesions, at the time of sample collection.
Analytical sensitivity and specificity of quantitative polymerase chain reaction
The quantitative polymerase chain reaction was performed as a confirmatory assay to determine M. leprae positive samples. The limit of detection for RLEP based quantitative polymerase chain reaction assay was 10 fg that is equivalent to approximately three M. leprae organisms13 and the standard curve showed reproducibility with a negative correlation between cycle threshold and DNA concentration of M. leprae [Figure 2a]. To evaluate the specificity of the quantitative polymerase chain reaction assay, control parasite DNA samples along with M. leprae genomic DNA were subjected to the quantitative polymerase chain reaction. The assay specifically detected M. leprae DNA. The cycle threshold value for all control samples was higher than the negative control (negative cutoff value of >34) indicating 100% specificity of the assay [Figure 2b].
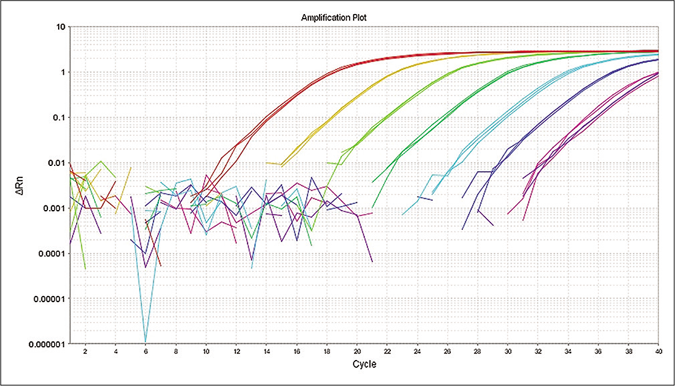
- Limit of detection for M. leprae at 10 fold dilution (1 ng/μL–1 fg/μL) by quantitative polymerase chain reaction
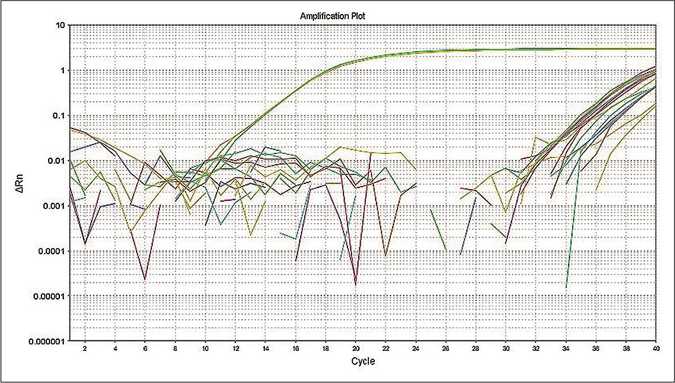
- Specificity of quantitative polymerase chain reaction primers for detection of M. leprae was tested using DNA from other organisms.
Analytical sensitivity and specificity of loop-mediated isothermal amplification assay
The loop-mediated isothermal amplification assay was performed using serially diluted M. leprae genomic DNA samples (tenfold; 1ng/μL to 1fg/μL). The analytical sensitivity of loop-mediated isothermal amplification in 60 min was 100 fg, equivalent to 30 organisms [Figure 3a]. To evaluate the specificity of the loop-mediated isothermal amplification assay, parasite DNA controls along with M. leprae genomic DNA were subjected to the loop-mediated isothermal amplification reaction. The assay detected M. leprae DNA as indicated by green color while all the control samples remained orange after completion of the reaction, suggesting that no amplification occurred (specificity- 100%; 95% confidence interval 92.9%–100%) [Figure 3b].
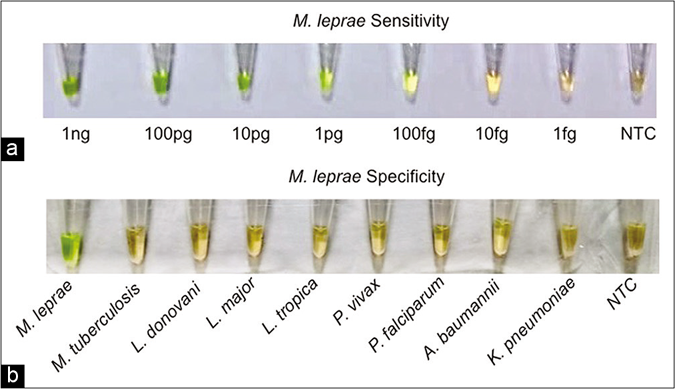
- (a) Visual detection of amplified loop-mediated isothermal amplification products using SYBR green I. Sensitivity for M. leprae (1 ng/ μL–1 fg/μL), (b) Visual detection of amplified loop-mediated isothermal amplification products using SYBR green I
Application of loop-mediated isothermal amplification assay in clinical samples
The loop-mediated isothermal amplification assay was applied to clinical samples and its efficacy was compared with the routinely used diagnostic tests (histopathology/microscopy) for M. leprae detection. Out of the 50 M. leprae positive tissue samples as confirmed by quantitative polymerase chain reaction, 47 tissue samples produced green color after 60 min incubation in loop-mediated isothermal amplification assay (sensitivity 94%; 95% confidence interval 83.5%–98.8%), a positive predictive value of 100% and negative predictive value of 93% (95% confidence interval 81.7%–97.6%) with an accuracy of 96.6% (95% confidence interval 90.5%–99.3%) with the assay being negative in all 25 other disease control samples, giving it a specificity of 100% (95% confidence interval 86.28% to 100.00%), while only 31 were positive for M. leprae in histopathology reports (sensitivity 62%; 95% confidence interval 47.2%–75.%) [Table 2 and Figure 4a]. Amongst these tissue samples, 37 out of 39 multibacillary samples (sensitivity 94.9%; 95% confidence interval 82.7%– 99.4%) and ten out of 11 paucibacillary samples (sensitivity 90.9%; 95% confidence interval 58.7%–99.8%) were positive by loop-mediated isothermal amplification assay whereas only 26 multibacillary samples (sensitivity 66.6%; 95% confidence interval 51%–79.4%) and five paucibacillary samples (sensitivity 45.5%; 95% confidence interval 21.3%– 72%) were found positive in histopathology reports [Table 2].
| Assays | Sensitivity %, 95% CI | |
|---|---|---|
| Multibacillary patients (n=39) | Paucibacillary patients (n=11) | |
| Tissue punch samples (n=50) | ||
| qPCR | 100% (n=39/39), 1-100% | 100% (n=11/11), 74-100% |
| LAMP | 94.9% (n=37/39), 82.7-99.4% | 90.9% (n=10/11), 58.7-99.8% |
| Histopathology | 66.6% (n=26/39), 51-79.4% | 45.5% (n=5/11), 21.3-72% |
| Slit aspirate samples (n=50) | ||
| qPCR | 87.2% (n=34/39), 72.6-95.7% | 72.7% (n=8/11), 39-94% |
| LAMP | 87.2% (n=34/39), 72.6-95.7% | 72.7% (n=8/11), 39-94% |
| Microscopy | 53.9% (n=21/39), 38.6-68.4% | 18.2% (n=2/11), 5.1-47.7% |
LAMP: loop-mediated isothermal amplification, CI: confidence interval, qPCR: quantitative polymerase chain reaction
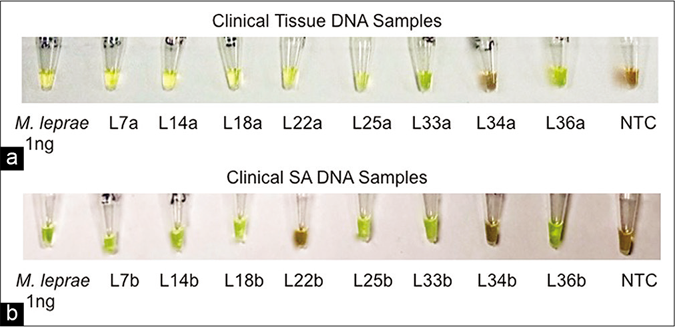
- (a) Visual detection of amplified loop-mediated isothermal amplification products in clinical samples. Tissue samples of the, (b) Visual detection of amplified loop-mediated isothermal amplification products in clinical samples. Slit aspirates samples
In the present study, we attempted to establish loop-mediated isothermal amplification assay for leprosy using slit aspirates of patients, as this sample collection procedure is far less invasive with advantages of ease of collection, storage and transportation. The slit aspirate samples of these patients showed 42/50 (sensitivity 84%; 95% confidence interval 70.9%– 92.8%) positives after 90 min incubation by loop-mediated isothermal amplification assay as well as quantitative polymerase chain reaction with the assay being negative in all 15 normal skin control samples, giving it a specificity of 100% (95% confidence interval 78.2% to 100.00%) and, a positive predictive value of 100% and negative predictive value of 83.3% (95% confidence interval 72.6%–90.4%) and an accuracy of 91% (95% confidence interval 83.2%–96.1%). Only 23/50 (sensitivity 46%; 95% confidence interval 31.8%–60.7%) slit aspirate samples were positive by microscopic observation [Table 2 and Figure 4b]. Amongst these slit aspirate samples, 34 out of 39 multibacillary samples (sensitivity 87.2%; 95% confidence interval 72.6%– 95.7%) and eight out of 11 paucibacillary samples (sensitivity 72.7%; 95% confidence interval 39%–94%) were positive by loop-mediated isothermal amplification assay whereas only 21 multibacillary samples (sensitivity 53.9%; 95% confidence interval 38.6%–68.4%) and only two paucibacillary samples (sensitivity 18.2%; 95% confidence interval 5.1%–47.7%) were found positive by microscopy [Table 2].
Discussion
Early diagnosis of leprosy is critical to control, along with its transmission and prompt treatment, as the nerve damage associated with the disease is permanent and irreversible. At present, diagnosis is based on clinical manifestations followed by the classical confirmatory tests such as histopathology or microscopy; however, these techniques suffer from low sensitivity. The cost-effective isothermal amplification-based techniques have emerged to substitute expensive molecular methods.18 Amongst these isothermal amplification methods, loop-mediated isothermal amplification fulfills all the requisites of the WHO-ASSURED criteria – affordable, sensitive, specific, user-friendly, robust and reliable, equipment-free and deliverable to those in need. Till date, loop-mediated isothermal amplification has been successfully applied in the diagnosis of various diseases such as leishmaniasis,22 tuberculosis,23 malaria,24 dengue,25 chikungunya,26 African trypanosomiasis27 and many more. Herein, we report a highly sensitive loop-mediated isothermal amplification assay specific for leprosy and its application on tissue and slit aspirate samples of leprosy patients. The loop-mediated isothermal amplification primers were designed against the RLEP gene, a highly conserved repetitive sequence with high copy number (n = 37).10 The developed assay was highly sensitive showing an analytical sensitivity of 100 fg that is equivalent to approximately 30 M. leprae organisms. An earlier attempt on developing leprosy specific loop-mediated isothermal amplification assay showed lower sensitivity (~50 M. leprae organism) without demonstrating its utility in clinical samples.28
The loop-mediated isothermal amplification assay for leprosy showed 100% specificity when tested on a range of organisms causing co-endemic diseases such as tuberculosis, leishmaniasis and malaria and other bacterial infections. In the present study, we used closed-tube technology that gave better sensitivity, clarity in the visualization of the results and minimized the risk of cross-contamination associated with conventional loop-mediated isothermal amplification thereby increasing its applicability in the field. The closed-tube loop-mediated isothermal amplification assays, either by using waxdye capsules29,30 or adding dye in tube cap at the beginning of reaction22,31 has been used widely to avoid cross-contamination. Our loop-mediated isothermal amplification assay for leprosy showed an overall sensitivity of 94% using tissue samples which were significantly higher than the routinely used histopathology (62%). The assay also showed a distinct advantage in detecting paucibacillary cases with 90.9% sensitivity in comparison with histopathology (45% sensitivity).
In our study, we attempted loop-mediated isothermal amplification assay using slit aspirates to minimize invasive procedure of sample collection. Skin slit aspirate is far less invasive than tissue biopsy as it does not require anesthesia, suturing, long healing time and leaves no mark after healing. Among different sampling techniques, slit aspirate sampling has the advantages of ease of collection, storage and transportation and most importantly can be effective in case-contact studies, a requisite to control leprosy transmission.32 The loop-mediated isothermal amplification assay was as effective as a quantitative polymerase chain reaction for detection of M. leprae (sensitivity 84%) using slit aspirate sample while being markedly superior to microscopy (sensitivity 46%). In fact, the loop-mediated isothermal amplification assay performed better than the reported polymerase chain reaction-based molecular tests that exhibited a sensitivity of approximately 76%.14,17 Furthermore, 8 out of 11 (72.7%) slit aspirates from paucibacillary patients tested positive for M. leprae as compared to two out of 11 (18.2%) by routine microscopy. Thus, based on the results observed in paucibacillary patients, our loop-mediated isothermal amplification assay can indeed be advantageous for detecting M. leprae in the early stage of the disease which could be crucial in controlling disease transmission.
Limitations
The paucibacillary patients constituted only 22% of the total cases enrolled in the study. Our loop-mediated isothermal amplification assay needs to be extended to a larger number of paucibacillary cases to establish its utility in early detection of the disease.
Conclusion
Loop-mediated isothermal amplification was shown to be a simple and rapid method for the laboratory identification of M. leprae in clinical samples. Our loop-mediated isothermal amplification assay has advantages over presently available diagnostics in terms of sensitivity and specificity and can undoubtedly be volunteered as an efficient tool for diagnosis of leprosy as it is simpler, faster, cost-effective and easy to perform with the potential of field applicability.
Acknowledgment
We wish to thank all the patients, nurses and clinicians from Safdarjung Hospital, New Delhi, as well as laboratory, research, and administrative staff, who took part in or contributed to this study.
Declaration of patient consent
The authors certify that they have obtained all appropriate patient consent.
Financial support and sponsorship
This research received grant from The Indian Council of Medical Research, New Delhi and The Science and Engineering Research Board (SERB), Department of Science and Technology, Government of India, in the form of SERB-National Post Doctoral Fellowship (Grant no.-PDF/2016/003200). PS is a JC Bose Fellow.
Conflicts of interest
There are no conflicts of interest.
References
- Genome-wide comparison of medieval and modern Mycobacterium leprae. Science. 2013;341:179-83.
- [CrossRef] [PubMed] [Google Scholar]
- Global leprosy update, 2016 Accelerating reduction of disease burden. Wkly Epidemiol Rec. 92:501-19.
- [Google Scholar]
- Current situation of leprosy in India and its future implications. Indian Dermatol Online J. 2018;9:83-9.
- [CrossRef] [PubMed] [Google Scholar]
- Diagnosing multibacillary leprosy: A comparative evaluation of diagnostic accuracy of slit-skin smear, bacterial index of granuloma and WHO operational classification. Indian J Dermatol Venereol Leprol. 2008;74:322-6.
- [CrossRef] [PubMed] [Google Scholar]
- Diagnostic accuracy of slit skin smears in leprosy. Pak Armed Forces Med J. 2015;65:649-52.
- [Google Scholar]
- Clinical and histopathological correlation in the classification of leprosy. Int J Lepr Other Mycobact Dis. 1993;61:433-8.
- [Google Scholar]
- Clinico histopathological correlation in leprosy. Dermatol Online J. 2012;18:2.
- [CrossRef] [PubMed] [Google Scholar]
- qPCR detection of Mycobacterium leprae in biopsies and slit skin smear of different leprosy clinical forms. Braz J Infect Dis. 2017;21:71-8.
- [CrossRef] [PubMed] [Google Scholar]
- PCR primers that can detect low levels of Mycobacterium leprae DNA. J Med Microbiol. 2001;50:177-82.
- [CrossRef] [PubMed] [Google Scholar]
- PCR-based techniques for leprosy diagnosis: From the laboratory to the clinic. PLoS Negl Trop Dis. 2014;8:e2655.
- [CrossRef] [PubMed] [Google Scholar]
- Enumeration of Mycobacterium leprae using real-time PCR. PLoS Negl Trop Dis. 2008;2:e328.
- [CrossRef] [PubMed] [Google Scholar]
- RLEP PCR as a definitive diagnostic test for leprosy from skin smear samples in childhood and adolescent leprosy. Indian J Lepr. 2016;88:193-7.
- [Google Scholar]
- Comparison of PCR mediated amplification of DNA and the classical methods for detection of Mycobacterium leprae in different types of clinical samples in leprosy patients and contacts. Lepr Rev. 2003;74:18-30.
- [CrossRef] [PubMed] [Google Scholar]
- Comparative evaluation of PCR amplification of RLEP, 16S rRNA, rpoT and Sod A gene targets for detection of M. leprae DNA from clinical and environmental samples. Int J Mycobacteriol. 2015;4:54-9.
- [CrossRef] [PubMed] [Google Scholar]
- Critical analysis: Use of polymerase chain reaction to diagnose leprosy. Braz J Pharm Sci. 2016;52:163-9.
- [CrossRef] [Google Scholar]
- Miniaturized isothermal nucleic acid amplification: A review. Lab Chip. 2011;11:1420-30.
- [CrossRef] [PubMed] [Google Scholar]
- Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63.
- [CrossRef] [PubMed] [Google Scholar]
- The direct boil-LAMP method: A simple and rapid diagnostic method for cutaneous leishmaniasis. Parasitol Int. 2014;63:785-9.
- [CrossRef] [PubMed] [Google Scholar]
- Detection of Fusarium graminearum DNA using a loop-mediated isothermal amplification (LAMP) assay. Int J Food Microbiol. 2010;140:183-91.
- [CrossRef] [PubMed] [Google Scholar]
- Development of a rapid loop-mediated isothermal amplification assay for diagnosis and assessment of cure of Leishmania infection. BMC Infect Dis. 2017;17:223.
- [CrossRef] [PubMed] [Google Scholar]
- Feasibility and operational performance of tuberculosis detection by loop-mediated isothermal amplification platform in decentralized settings: Results from a multicenter study. J Clin Microbiol. 2016;54:1984-91.
- [CrossRef] [PubMed] [Google Scholar]
- Rapid detection of Plasmodium vivax in saliva and blood using loop mediated isothermal amplification (LAMP) assay. J Infect. 2013;67:245-7.
- [CrossRef] [PubMed] [Google Scholar]
- Development and validation of four one-step real-time RT-LAMP assays for specific detection of each dengue virus serotype. PLoS Negl Trop Dis. 2018;12:e0006381.
- [CrossRef] [PubMed] [Google Scholar]
- Development of a single-tube one-step RT-LAMP assay to detect the Chikungunya virus genome. PLoS Negl Trop Dis. 2018;12:e0006448.
- [CrossRef] [PubMed] [Google Scholar]
- Loop-mediated isothermal amplification (LAMP) method for rapid detection of Trypanosoma brucei rhodesiense. PLoS Negl Trop Dis. 2008;2:e147.
- [CrossRef] [PubMed] [Google Scholar]
- Development of rapid and simple genomic diagnostic method. Nihon Hansenbyo Gakkai Zasshi. 2006;75:265-9.
- [CrossRef] [PubMed] [Google Scholar]
- A closed-tube detection of loop-mediated isothermal amplification (LAMP) products using a wax-sealed fluorescent intercalator. J Nanosci Nanotechnol. 2013;13:3999-4005.
- [CrossRef] [PubMed] [Google Scholar]
- New closed tube loop mediated isothermal amplification assay for prevention of product cross-contamination. MethodsX. 2014;1:137-43.
- [CrossRef] [PubMed] [Google Scholar]
- Validation of SYBR green I based closed tube loop mediated isothermal amplification (LAMP) assay and simplified direct-blood-lysis (DBL)-LAMP assay for diagnosis of visceral leishmaniasis (VL) PLoS Negl Trop Dis. 2018;12:e0006922.
- [CrossRef] [PubMed] [Google Scholar]
- Reliable diagnosis of post-kala-azar dermal leishmaniasis (PKDL) using slit aspirate specimen to avoid invasive sampling procedures. Trop Med Int Health. 2013;18:268-75.
- [CrossRef] [PubMed] [Google Scholar]






