Translate this page into:
Effects of keratinocyte-derived and fibroblast-derived exosomes on human epidermal melanocytes
Corresponding author: Dr. Ru-zhi Zhang, Department of Dermatology, The Third Affiliated Hospital of Soochow University, Changzhou, China. zhangruzhi628@163.com
-
Received: ,
Accepted: ,
How to cite this article: Shi HX, Zhang RZ, Xiao L, Wang L. Effects of keratinocyte-derived and fibroblast-derived exosomes on human epidermal melanocytes. Indian J Dermatol Venereol Leprol 2022;88:322-31.
Abstract
Background:
Exosomes have been demonstrated to carry proteins, membrane lipids, mRNAs and microRNAs which can be transferred to surrounding cells and regulate the functions of those recipient cells.
Objectives:
The objective of the study was to investigate the effects of exosomes released by keratinocytes and fibroblasts on the proliferation, tyrosinase activity and melanogenesis of melanocytes.
Methods:
Melanocytes, keratinocytes and fibroblasts obtained from human foreskin were cultured and exosomes secreted by keratinocytes and fibroblasts were harvested from the culture supernatants by ultracentrifugation. Each exosome fraction was divided into two parts; one part was subjected to high-throughput sequencing using an Illumina HiSeq sequencer to characterize the microRNA expression profiles, while the other part was labeled with the fluorescent dye PKH67 and was then co-cultivated with epidermal melanocytes.
Results:
High-throughput sequencing analysis showed 168 differentially expressed microRNA within exosomes derived from keratinocytes and from fibroblasts, 97 of those being up-regulated with the other 71 down-regulated. Gene ontology analysis showed that the target genes responsible for these differentially expressed microRNAs were mainly enriched in the protein-binding region of molecular functions. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis showed that target genes regulated by differentially expressed microRNA were mainly involved in mitogen-activated protein kinase (MAPK) signaling pathway, Ras signaling pathway, cAMP signaling pathway and Wnt signaling pathway. Keratinocyte-derived exosomes were taken up by melanocytes co-cultured with them and promoted the proliferation, tyrosinase activity and melanin synthesis of those melanocytes. However, fibroblast-derived exosomes had no similar effects on melanocytes.
Conclusion:
Keratinocyte-derived exosomes but not fibroblast-derived exosomes were taken up by melanocytes in co-culture and significantly stimulated their proliferation, tyrosinase activity and melanin synthesis. Those different effects may be mainly due to the differential expression of microRNAs in exosomes derived from the different types of cells.
Limitations:
Electron microscopy of the obtained exosomes and in-depth study of apparently differentially expressed microRNAs were not performed.
Keywords
Exosomes
fibroblasts
high-throughput sequencing
keratinocytes
melanocytes
microRNAs
Plain Language Summary
Exosomes are extracellular vesicles originating from endosomes that can carry protein, mRNAs, microRNAs and cytokines. Our study has shown that exosomes released by keratinocytes may target co-cultured melanocytes. The biological activities of these melanocytes are affected, such as tyrosinase activity, cell proliferation, melanogenesis, etc. High-throughput sequencing showed differentially expressed microRNA within melanocytes derived from keratinocytes and fibroblasts, which may be the cause of these effects.
Introduction
Exosomes are secreted and released by cells as cup-shaped lipid-bilayer membrane-limited vesicles 50–100 nm in diameter.1 Previous studies have addressed the functions of extracellular vesicles in physiological and pathological conditions based on their biological activities and molecular compositions. Recently, the role of exosomes has been highlighted in research studies of intercellular communications and they have emerged as an important route of information exchange among cells involved in many physiological and pathological processes.2,3
During the biosynthesis of exosomes, many types of biomolecules derived from parental cells are encapsulated within them, including DNA, microRNAs, proteins and lipids that can be functional and can be delivered into other cells resulting in molecular signaling processes such as immune responses and oncogenesis.4 Many microRNAs have been shown to play critical roles in skin biology and the hair cycle. Recently, Lo et al. reported that exosomes released by keratinocytes carry microRNAs that are targeted to melanocytes and modulate the pigmented status of melanocytes by altering gene expression patterns and enzyme activities.5 Peter et al. attempted to identify microRNAs that interfered with the pigmentary process and to assess their functional roles and found 16 differentially expressed microRNAs, including a 15-fold differential expression of miR-145.6 Another report indicated that keratinocytes produce miR- 330-5p,7 which down-regulated the expression of TYR that plays a key role in pigmentation, miR-21a-5p,8 which targets SOX5 through the MITF pathway and miR-27a-3p,9 which regulates Wnt3a to modulate melanogenesis.
Melanogenesis directly affects the color of skin and hair. The normal synthesis and production of melanin protects the skin from damage caused by ultraviolet radiation.10 Melanocytes synthesize melanin within melanosomes and distribute mature melanosomes to surrounding keratinocytes through their dendrites, forming distinct epidermal melanin units. Keratinocytes not only passively accept melanosomes transferred by melanocytes but also affect the properties of melanocytes by secreting a series of cytokines which may be carried through exosomes.11
Based on these facts, we chose exosomes from epidermal keratinocytes and from dermal fibroblasts derived from human foreskins as targets and used a co-culture system to investigate the effects of those cell-derived exosomes on melanocyte functions. This study provides further evidence for factors that affect melanogenesis in human epidermal melanocytes.
Materials and Methods
Culture of epidermal melanocytes and keratinocytes and dermal fibroblasts
This study was approved by the ethics committee of the third affiliated hospital of Soochow University and was undertaken on the basis of the basic principles of the declaration of Helsinki. Written informed consent was obtained from patients.
Skin specimens were obtained from adult circumcisions and were immediately immersed in iodine solution for five min. After washing several times with phosphate-buffered saline (PBS) supplemented with penicillin, 400 U/mL and streptomycin, 400 μg/mL, subcutaneous adipose tissue was carefully removed using ophthalmic scissors. The remaining epidermal and dermal tissue were washed three times with PBS and then was manually cut into small pieces of 2 × 2 mm, which were then immersed in 0.25% Dispase-II solution for 2 h at 37°C with 5% CO2. Subsequently, the epidermis was separated from the dermis with the help of a pair of fine scissors and was trypsinized with a solution of 0.25% trypsin/0.02% ethylenediamine tetraacetic acid for 15 min to produce a single cell suspension which was filtered through a 200 μm membrane and then centrifuged at 1000 r/min for three min at room temperature. The cell pellet obtained was re-suspended with M254 medium (supplemented with human melanocyte growth supplement) or EpiLife medium (supplemented with human keratinocyte growth supplement), respectively, to promote the growth of melanocytes and keratinocytes. The dermal tissue was cut into pieces of 2 × 2 mm which were collected and immersed in a solution containing 0.25% type IV collagenase for 2 h and then in a solution of 0.25% trypsin/0.02% ethylenediamine tetraacetic acid for 20 min. After having successively undergone filtration and centrifugation, cell pellets were obtained. For fibroblast culture, the cells were suspended in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% FBS, 100U/ml penicillin and 0.1mg/ml streptomycin. In these experiments, melanocytes, keratinocytes and fibroblasts at fifth passages were used.
Preparation of conditioned medium
Keratinocytes and fibroblasts were seeded in 175 cm2 flasks and were maintained in the appropriate growth media which were changed once after complete cell adhesion. When 70% confluence of the cells was reached, they were transferred to serum-free Dulbecco’s modified Eagle medium containing antibiotic-antimycotic for 48 h. The conditioned media were harvested and centrifuged at 4°C, 2000 r/min at room temperature for 10 min to remove obvious cell debris. They were then filtered through a 0.2 μm membrane, aliquoted and stored at 4°C or −80°C before being used for the experiments described below. Fresh serum-free growth media were utilized as controls.
Isolation and purification of exosomes
Extraction of exosomes by ultracentrifugation
The conditioned media stored at –80°C were first thawed at 4°C, after which they were centrifuged at 3000 g for 15 min at 4°C to remove residual cells. The collected supernatants were further centrifuged at 10000 g for 30 min at 4°C to remove any cells or organelle fragments that may be present. Finally, the harvested supernatants were centrifuged at 100,000 g for 70 min at 4°C and pellets at the bottom of the ultracentrifuge tubes were used as the crude exosome fraction. To increase the purity of exosomes, they were suspended with Dulbecco’s phosphate-buffered saline and centrifuged again at 100,000 g for 70 min at 4°C. The resulting pellets were suspended in100 μl Dulbecco’s phosphate-buffered saline and stored at –80°C for subsequent co-culture experiments.
Extraction of exosomes by an exoEasy Maxi kit
Since the exoEasy Maxi kit method (QIAGEN, Germany) is only suitable for extracting exosomes from a small amount of supernatant, we concentrated the medium using ultrafiltration, after which the exosomes were extracted according to the instructions of the exoEasy Maxi kit. The extracted exosomes were suspended with 1 ml phosphate buffer saline and stored at –80°C for subsequent high-throughput sequencing assays.
miRNA array profiling
Exosomes extracted with the exoEasy Maxi kit were characterized using an Illumina HiSeq high-throughput sequencer. Total RNA, including microRNAs, was extracted from samples derived from keratinocytes and fibroblasts according to the manufacturer’s recommendations. microRNAs were isolated from total RNAs using gel size selection. Isolated microRNA fragments were subjected to a series of treatments including reverse transcription to form a cDNA library and linkers were ligated to both ends of each fragment and amplified into a miRNA library by RT-PCR. The quality of cDNA was measured by an Agilent 2100 Bioanalyzer profile. The obtained RNA library was deeply sequenced using an Illumina Hiseq™ 2000 sequencer (Shanghai OE Biotech. Co., Ltd.) and the sequences obtained were compared with the Rfam, cDNA sequence, species repeat library and miRBase to annotate the microRNAs detected.
Labeling exosomes with the fluorescent dyePKH67
To examine whether exosomes from keratinocytes and/or fibroblasts can be taken up by melanocytes, the exosome fractions were labeled with the fluorescent dye PKH67 according to the manufacturer’s directions. Briefly, 100 μL of each exosome suspension were diluted in sterile PBS to 250 μL. The PKH67 dye (1.0 μL) was diluted in 250 μL Diluent C (PKH67 solution) and 250 μL PKH67 solution and 250 μL diluted exosomes were mixed in a centrifugation tube and incubated at room temperature for five min. An equal volume of 1% bovine serum albumin was added to bind the excess PKH67 dye. The PKH67-stained exosomes were washed three times in sterile PBS to remove excess dye and were then collected by centrifugation at 120,000 g for 70 min. Finally, the PKH67-labeled exosomes were re-suspended in Dulbecco’s modified eagle medium and then co-cultured with epidermal melanocytes at 37°C and 5% CO2 for 48 h.
Tyrosine (TYR) activity assay
TYR activity was determined in microtiter plates as previously described with slight modifications. Melanocytes were treated with exosomes from keratinocytes (incubated with exosomes from an equal number of keratinocytes) for 24 h. The cell supernatants were then replaced with 90 μL phosphate buffer containing 1% Triton X-100. The microtiter plates were placed in a –80°C refrigerator for 30 min and the cells were lysed completely. After vortexing, the cells were clarified by centrifugation at 13,000 r/min for 10 min at 4°C. Ten microliters of the TYR substrate L-Dopa (15 mM) was incubated with 90 μL of each extract in a 96-well plate for 2 h at 37°C. Absorbance values were then detected at 475 nm using a Multiscan Spectrum microplate reader (DNM-9602).
Melanin assay
The melanocytes were treated with exosomes from keratinocytes or fibroblasts (incubated with exosomes or medium isolated from an equal number of cells) for 48 h and were then disrupted by sonication in 50 mM Tris-HCl, pH7.4, 2mM ethylenediamine tetraacetic acid, 150 mM NaCl, 1 mM dithiothreitol and protease inhibitors. Pigment was pelleted at 20,000 g for 15min at 4°C, rinsed once in ethanol/ether (1:1) and dissolved in 2M NaOH/20% dimethyl sulfoxide at 60°C. Melanin contents were measured as OD at 492 nm (DNM-9602).
Results
Preliminary analysis of sequencing results
After sequencing using an Illumina HiSeq sequencer, 21,275,528 and 29,532,598 raw data before deadaptor were obtained from exosomes derived from keratinocytes and from fibroblasts, respectively. The obtained reads were preliminarily filtered through the processes of removing adaptors at both ends of the sequence, removing low quality reads and decontaminating to retain clean reads. The clean reads of 15–41 nt for analysis of the two samples were 18,889,261 and 25,046,185, respectively. After the redundancy, the unique clean reads were 195,360 and 293,671 [Table 1].
| Sample | Raw before deadaptor | Raw reads | Reads trimmed length | Reads trimmed Q20 | Reads trimmed N | Clean reads | Clean reads unique |
|---|---|---|---|---|---|---|---|
| KC-derived exosomes | 21475982 | 21464215 | 18990124 | 18912197 | 18889261 | 18889261 | 195360 |
| FB-derived exosomes | 25932598 | 29528664 | 25179020 | 25077460 | 25046185 | 25046185 | 293671 |
KC-derived: Keratinocyte-derived; FB-derived: Fibroblast-derived
Length distribution of clean reads in two samples
The length distribution of clean reads helps to compare microRNAs of the two different processed samples. In general, the length distribution in the sample peaks at 21–25 bp and sometimes there was another peak that was around 31–33 bp, which may reflect the distribution of piRNAs, tRNAs and/ or rRNAs. In some cases, such as in samples infected with a virus or treated with a drug, there can be abnormalities in the length distribution. Therefore, the length distribution of a sample is a good criterion for assessing the sample condition and for assessing the biological state of the sample. The length of reads in exosomes derived from keratinocytes was mainly distributed between 30 and 33bp and peaked at 31 and 32 bp. The length of reads in exosomes derived from fibroblasts was mainly distributed between 29 and 32 bp and peaked at 31 bp. This indicates that the distribution of reads in exosomes released by keratinocytes and fibroblasts has an obvious similarity.
Comparison and annotation of clean reads
There are many kinds of microRNAs, including miRNA, tRNA (tiRNA and tRFs), rRNA, piRNA and snoRNA. We aligned the clean reads obtained above with the Rfam databases to remove rRNA, scRNA, snoRNA, snRNA, tRNA as much as possible. There was no mismatch between these filtered sequences and transcripts. Reads with a length greater than 26 bp that completely aligned with the transcript were considered to be degraded fragments of microRNAs. The results showed that there were not many microRNA reads and among those reads, un-annotated reads accounted for the majority. The known microRNAs in the two samples accounted for 0.6% and 0.14%, respectively [Figures 1a and 1b]. Subsequently, they were aligned with the repeat databases to identify possible repeat reads and to filter them out. Finally, the aligned reads were mismatched with the mature microRNA reads in miRBase and the aligned reads were considered to be known microRNAs. The number of screened microRNAs and their reads is shown in Table 2 and 296 and 294 microRNAs were detected in the two samples, respectively.
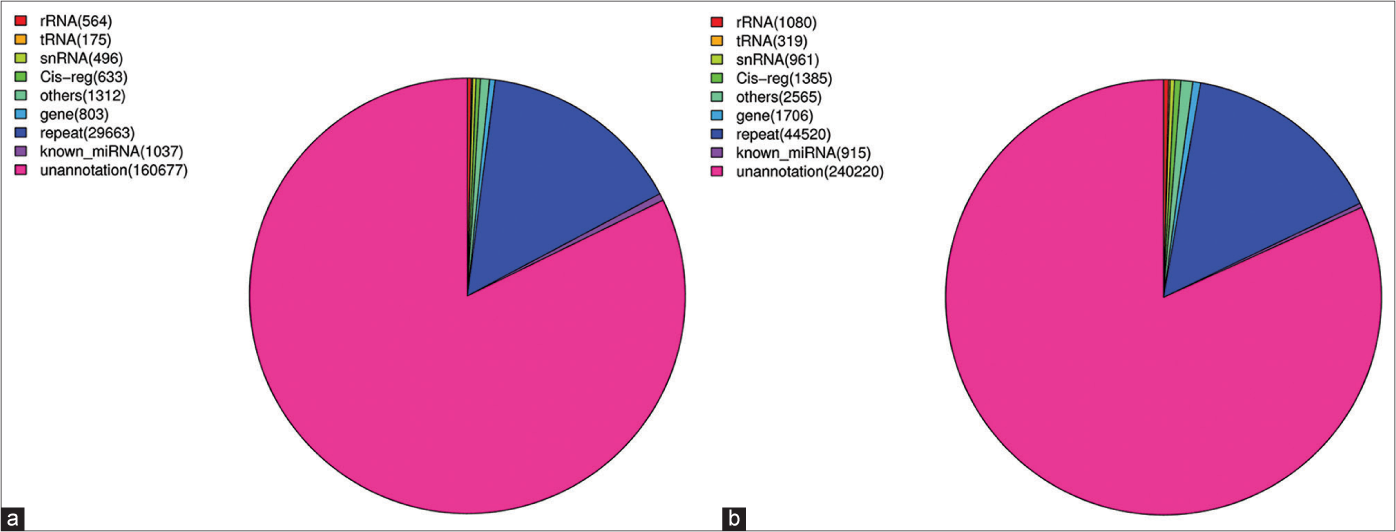
- Distribution of miRNAs and other RNAs in KC-derived exosomes (a) (Sample A) and FB-derived exosomes (b) (Sample B). The known miRNAs in those two samples accounted for 0.6% and 0.14%, respectively
| KC-derived exosomes | FB-derived exosomes | |
|---|---|---|
| Reads | 18889261 | 25046185 |
| Aligned reads | 114166 | 35632 |
| Known miRNA categories | 296 | 294 |
First base composition
The process of microRNA development from a precursor to a mature body is digested by Dicer. In general, the specificity of the restriction enzyme cutting site makes the first base of the mature microRNAs strongly biased toward the base U. Therefore, we analyzed the first base of the 5’- end of mature microRNAs detected in each sample. The results showed that the first base of the 5’- end of microRNAs of Samples A and B (15–25 nt) was mainly base U [Figures 2a and 2b]. This also shows that our sequencing quality is sufficient.

- First base of the 5’-end of miRNAs. The first base of the 5’-end of miRNAs of KC-derived exosomes (a) (Sample A) and FB-derived exosomes (b) (Sample B) (15–25 nt) was mainly base U
Screening of differentially expressed miRNA in exosomes derived from keratinocytes and from fibroblasts
Some microRNAs are expressed both in keratinocyte-derived and fibroblast-derived exosomes, while some are only expressed in one sample or the other. The expression level of microRNAs is usually determined by its abundance; the higher the abundance of microRNAs, the higher the expression level. To compare the expression of microRNAs in the two samples, we standardized their expression in the samples to transcript per million (TPM) and the p value was calculated using the Audic Claverie formula (P-value reflects the importance of differentially expressed microRNAs among samples; the smaller the P-value, the more significant the difference). microRNAs screened by P < 0.05 and TPM difference multiples >2 are the differentially expressed microRNAs in keratinocyte-derived exosomes and fibroblast-derived exosomes. The results show that there were 168 known microRNAs with significant differences in expression, of which 97 microRNAs were upregulated in keratinocyte-derived exosomes while 71 miRNAs were downregulated in fibroblast-derived exosomes. Clustering analysis of the data was performed using Cluster 3.0 software [Figure 3].
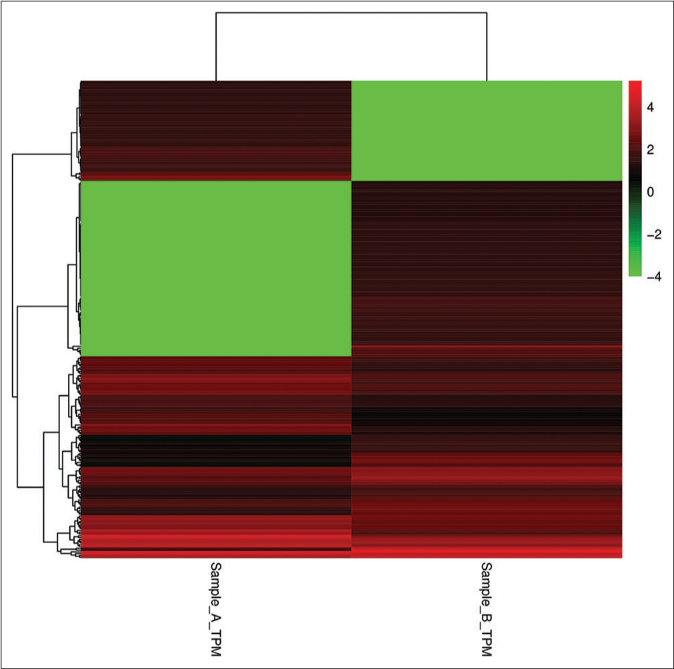
- Clustering analysis of the data of KC-derived exosomes (Sample A) and FB-derived exosomes (Sample B). The abscissa represents sample clustering and each column represents one sample. The clustering is based on the similarity of gene expression between samples. The closer the gene expression between samples is, the closer it is. The ordinate represents gene clustering, each row represents one gene and clustering is based on the similarity of gene expression in the sample and the closer the gene is expressed in the sample, the closer it is. The color scale represents the abundance of gene expression. The deeper the red color, the greater expression was upregulated, the deeper the green color and the greater expression was down-regulated
Biological function analysis of differentially expressed miRNAs
Gene ontology enrichment analysis of differentially expressed exosomal microRNAs standardizes gene products according to their functions, biological pathways and cell localization, that is, simple annotation of gene products. Through gene ontology enrichment analysis, we can roughly understand the biological functions, pathways and cell localizations of differential gene enrichment. The three classifications of gene ontology describe the biological processes, molecular functions and cellular components of genes. Ontology analysis of these target genes revealed that they could be annotated in 16,167 gene ontology terms and we screened the top 10 gene ontology terms in the three categories. In the biological processes, the target genes were mainly enriched in the transcription, DNA-templated, regulation of transcription, DNA-templated and signal transduction. In the cellular component, the target genes were mainly enriched in the cytoplasm, nucleus, plasma membrane and cytosol. In the molecular function, the target genes were mainly enriched in protein binding.
Most significantly enriched gene ontology terms are rooted in the regulatory process. KEGG metabolic pathway analysis can reveal the metabolic pathways that are significantly altered by differentially expressed microRNAs. These target genes can be annotated into 302 metabolic pathways. We screened the top 20 signal pathways for enrichment and found that the differentially expressed miRNA-regulated target genes are mainly involved in the MAPK signal transduction pathway, Ras signaling pathway, Rap1 signaling pathway, cAMP signaling pathway, endocytosis and axon guidance, Wnt signaling pathway, actin cytoskeleton regulation and neurotrophin signal transduction pathway.
Internalization of exosomes by melanocytes and morphological observations
Melanocytes were randomly allocated into a blank group (cultured melanocytes only), an experimental Group 1 (melanocytes co-cultured with exosomes secreted by fibroblasts) and an experimental Group 2 (melanocytes co-cultured with exosomes secreted by keratinocytes). These exosomes had been labeled by the fluorescent dye PKH67. We found that there were bright green fluorescent particles in melanocytes of Group 2, which were mainly distributed in the cell bodies and dendrites of melanocytes [Figures 4a and 4b], but these were not observed in Group 1, indicating that exosomes secreted by keratinocytes are transported into melanocytes by some mechanism, whereas exosomes secreted by fibroblasts do not enter melanocytes.

- Characteristics of the internalization of exosomes in melanocytes. (a) Bright green fluorescent particles were observed in melanocytes co-cultured with keratinocyte-derived exosomes, which were mainly distributed in their cell bodies and dendrites
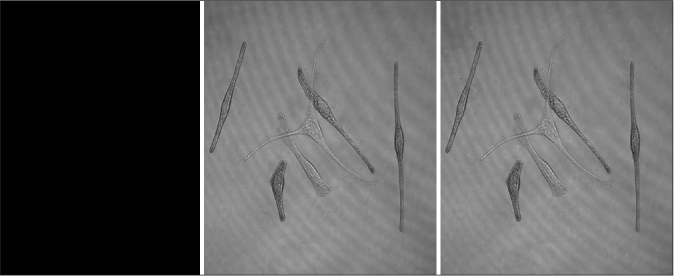
- Characteristics of the internalization of exosomes in melanocytes. No bright green fluorescent particles were observed in melanocytes co-cultured with fibroblast-derived exosomes (CarlZeiss, LSM710)
After 48 h of incubation, the proliferation and morphological characteristics of those three groups of melanocytes were observed using a light microscope. The results showed that there was no significant difference in melanocyte density between the blank group and the Group 1 at low magnification [Figurces 5a and 5b], but melanocytes of the Group 2 had proliferated rapidly, were densely distributed and were arranged very closely [Figure 5c]. They all have the typical morphological characteristics of melanocytes. Melanocytes in the blank group and in Group 1b were similar in morphology, showing bipolar or tripolar dendrites [Figures 5d and 5e]. However, melanocytes in Group 2 showed multiple dendrites, had a tendency to grow in clusters and the dendrites did not stretch out as in the other 2 groups [Figure 5f].

- Morphological observations of melanocytes. (a, b) The density of melanocytes in the blank group and the experimental Group 1 (melanocytes co-cultured with exosomes secreted by fibroblasts) was similar (×40). (c) Melanocytes in the experimental Group 2 (melanocytes co-cultured with exosomes secreted by keratinocytes) proliferated rapidly and were distributed densely (×40). (d-f) The three groups of cells were all typical melanocytes and the melanocytes in the blank group and in the experimental Group 1 were similar in morphology, showing bipolar or tripolar dendrites (d and e). (f) However, melanocytes in the experimental Group 2 showed multiple dendrites, had a tendency to grow in clusters and the dendrites did not stretch as much as in the experimental Group 1 and the blank group (×100) (OLYMPUS, CKX41-A32RC)
Tyrosinase activity of the three groups of melanocytes
The tyrosinase activity of the three groups of melanocytes was then measured. The results revealed that keratinocyte-derived exosomes had a significant stimulatory effect on tyrosinase activity compared with the blank group (P < 0.001) and Group 1 (P < 0.001). There was no significant difference between Group 1 and the blank group (P > 0.05) [Figure 6].
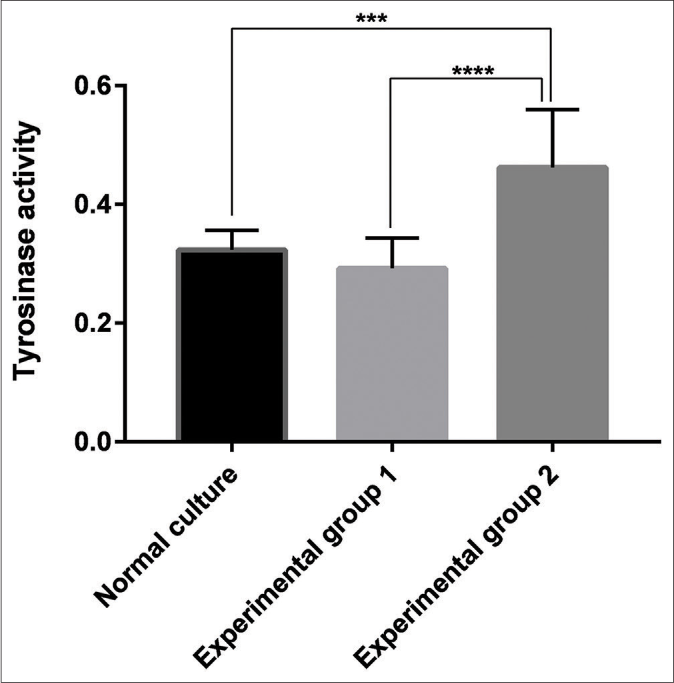
- Tyrosinase activity of the blank group, the experimental Group 1 (melanocytes co-cultured with exosomes secreted by fibroblasts) and the experimental Group 2 (melanocytes co-cultured with exosomes secreted by keratinocytes). Keratinocyte-derived exosomes had a significant stimulatory effect on tyrosinase activity compared with the blank group (P < 0.001) and the experimental Group 1 (P < 0.001). There was no significant difference between the experimental Group 1 and the blank group (P > 0.05)
Melanin content of the three groups of melanocytes
Measurement of the melanin content of the three groups of melanocytes demonstrated that keratinocyte-derived exosomes had a significant stimulatory effect on melanin content compared with the blank group (P = 0.0021) and Group 1 (P < 0.001). However, no significant difference between Group 1 and the blank group was observed (P > 0.05) [Figure 7].
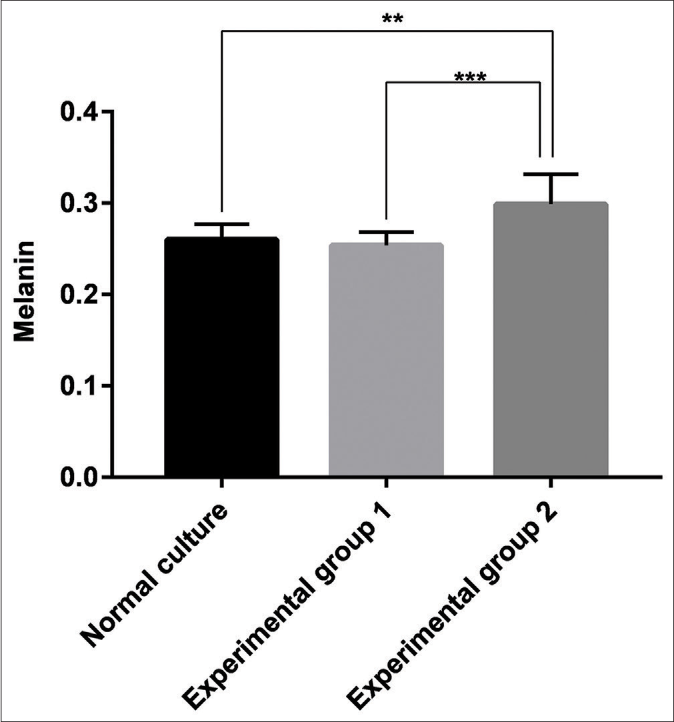
- Melanin content of the blank group, the experimental Group 1 (melanocytes co-cultured with exosomes secreted by fibroblasts) and the experimental Group 2 (melanocytes co-cultured with exosomes secreted by keratinocytes). Keratinocyte-derived exosomes had a significant stimulatory effect on melanin content compared with the blank group (P < 0.05, P = 0.0021) and the experimental Group 1 (P < 0.001). However, no significant difference between the control group and the blank group was observed (P > 0.05)
Discussion
It is widely known that melanogenesis is a tightly regulated process. Melanogenesis is the result of co-action of many types of cells in the skin, such as melanocytes, keratinocytes, fibroblasts and immune cells. These cells are also actively involved in the regulation of melanocyte behavior through the production of paracrine factors. Indeed, previous papers showed that melanogenesis is induced by ultraviolet B irradiated keratinocytes or directly by keratinocytes from high phototype individuals. Ultraviolet radiation is the main stimulus for skin pigmentation; in vivo, it increases proliferation and melanization of melanocytes. To stimulate melanogenesis, ultraviolet radiation can act directly on cultured melanocytes or indirectly through the release of keratinocyte-derived factors. Production of basic fibroblast growth factor, endothelin-1 and GMCSF by keratinocytes is upregulated after ultraviolet light exposure and these peptides stimulate melanocyte growth.12
Besides keratinocytes, dermal fibroblast-derived soluble factors have been shown to play important roles in the development of constitutive human skin color and the regulation of melanogenesis under facultative conditions (ultraviolet irradiation). These soluble factors include:
(1) Pleiotrophin (PTN), that is expressed in human skin cells, and in especially in high levels in fibroblasts. It inhibits melanogenesis through MITF degradation through the extracellular signal-regulated kinase 1/2 pathway in normal human melanocytes;13 (2) The KGF/KGF receptor system that has been shown to stimulate melanin production both in pigmented epidermal equivalents and human skin explants;14 (3) The fibroblast growth factor family member keratinocyte growth factor/ fibroblast growth factor 7 (KGF/FGF7) that induces melanosomes;15 and (4) DKK1 that has an inhibitory effect on MITF expression which results mainly from the decreased activity of GSK-3b and b-catenin. In addition, DKK1 upregulates the expression of myoactive tetradeca peptide which reduce TYR activity. Therefore, DKK1 acts on melanocytes by suppressing proliferation and melanin production.16
Exosomes are small extracellular vesicles (EVs) derived from multi-vesicular bodies or late endosomes that participate in intercellular communication.17 It is widely accepted that exosomes facilitate the direct extracellular transfer of proteins, lipids and RNAs, in vitro and in vivo, which enables them to participate in intercellular communication without direct contact.18 A large number of miRNAs carried by exosomes can participate in intercellular communication and miRNAs that selectively target melanocytes can regulate melanocyte pigmentation by changing gene expression patterns and/or enzyme activity.5 Studies have shown that miR-145,6 miR-675,19 miR-340,20 miR-218,21 miR-330-5p,7 miR-211,22 miR-27a-3p,9 miR-25,23 miR-155,24 miR-21a-5p8 and other miRNAs affect melanogenesis through various mechanisms.
Previous studies indicate that microRNAs regulate melanogenesis mainly through the following pathways: (1) some microRNAs targetted MITF and consequently regulated mRNA levels of TYR and TYRP1, such as miR-25;25 (2) miR-145 targetted Myo5a as a regulator in pigmentation in mouse and human melanocytes;6 (3) miR-21-5p targeted some proteins involved in melanogenesis pathway such as SOX5, beta-catenin, cyclin-dependent kinase 2 and MITF;26 (4) MicroRNA-141-3p and microRNA-200a-3p regulated α-melanocyte stimulating hormone-stimulated melanogenesis by directly targeting MITF;27 (5) miR-143-5p played a functional role in regulating alpaca melanocyte migration, proliferation and melanogenesis through direct targeting of TAK1;28 (6) miR-380-3p targeted SOX6 to regulate melanogenesis by influencing β-catenin and MITF transcription and translation, which reduced the expression of downstream genes, including TYR, TYRP1 and DCT;29 (7) miR-203 can reduce melanosome transport and promote melanogenesis by targeting KIF5B and through negative regulation of the CREB1/MITF/Rab27a pathway;30 (8) miR-27-3p inhibited melanogenesis by repressing Wnt3a protein expression;9 and (9) miR-125b regulated SRC homology 3 domain-binding protein 4 as a pigmentation-related gene.31
In our study, we performed microRNA array analysis on exosomes produced by keratinocytes and by fibroblasts for the first time. The results showed 168 known microRNAs with significantly different expression levels in both groups of exosomes, of which 97 were upregulated and 71 were downregulated.
The keratinocytes, melanocytes and fibroblasts used in this study were derived from human skin. Gene Ontology enrichment analysis revealed that the target genes of these differentially expressed miRNAs were mainly enriched in protein binding, metal ion binding, transcription, transcriptional regulation, cell components and other terms, all of which are closely related to cell growth and metabolism. KEGG pathway analysis showed that the target genes regulated by the differentially expressed miRNAs are mainly involved in the MAPK signaling pathway, Ras signaling pathway, Rap1 signaling pathway, cAMP signaling pathway, endocytosis, axon guidance, Wnt signaling pathway and regulation of actin cytoskeleton, neurotrophin signaling pathway and pathways in cancer. Therefore, the differentially expressed microRNAs identified in this study may target one or more genes and regulate one or more signaling pathways that affect the biological activities of melanocytes. Taking miR-211 (up-regulated) and miR-125b (down-regulated) as examples, we found that the differential expression multiple was 15 in keratinocyte- and in fibroblast-derived exosomes. The study by Sahoo et al. has established that miR-211and its target genes may play important roles in cellular metabolism in vitiligo cells and have therapeutic and biomarker potential.32 Similarly, a research study by Spiegelman and Elcheva revealed that miR-211 was significantly downregulated in biopsies from patients with vitiligo.33 Moreover, the loss of miR-211 expression correlated with the activity of the disease in nonlesional, perilesional and lesional skin. The data generated from small RNA deep sequencing showed that the miR-125b was significantly downregulated in keratinocyte-derived exosomes. Kyu-Han et al. demonstrated that the increased miR-125b expression inhibited pigmentary gene expression and consequently induced hypopigmentation.34 Therefore, we hypothesized that a study of the regulatory pathways between the expressions of these apparently differentially expressed microRNAs, their downstream targets and external signals, can reveal the pathogenesis of some pigmentary skin diseases and provide a promising new approach to therapy.
To investigate the effects of exosomes produced by different cell sources on melanocytes more intuitively, we observed the internalization process of exosomes, then determined tyrosinase activity and melanin content. PKH67, a common exosome marker that exhibits green fluorescence, was used to label exosomes. After co-culture for 48 h, some scattered green fluorescent particles could be seen in melanocytes co-cultured with keratinocyte-derived exosomes. However, this phenomenon did not appear in melanocytes co-cultured with fibroblast-derived exosomes. These findings indicated that exosomes secreted by keratinocytes are transported into melanocytes by a certain route, while those derived from fibroblasts are not internalized into melanocytes. Furthermore, we found that melanocytes treated with keratinocyte-derived exosomes proliferated rapidly and increased their dendricity, with an average of 3–4 branches. One-way ANOVA analysis showed that the tyrosinase activity and melanin content in the three groups were significantly different. In other words, keratinocyte-derived exosomes not only promoted the proliferation and dendricity of melanocytes but also enhanced their tyrosinase activity and production of melanin. However, fibroblast-derived exosomes did not play such roles and did not even enter melanocytes. This is an interesting phenomenon and we have carefully searched the literature, but no related studies have been reported.
Although there have been a few previous studies on the effects of exosomes released by keratinocytes on melanocytes, the novel aspect of this study was the introduction of exosomes secreted by fibroblasts. In addition, with the help of specific fluorescent markers, we attempted to observe the transport of exosomes derived from different types of cells using a confocal microscope. With high-throughput sequencing analysis, many differentially expressed microRNAs were found in exosomes released by keratinocytes and by fibroblasts, which provides a basis for further study of the effects of microRNAs on melanogenesis and the exploration of new therapies for the treatment of pigmentary skin diseases. In the future, in-depth studies of these clearly differentially expressed microRNAs, their target genes and the signaling pathways involved, may help to uncover the pathogenesis of pigmentary skin diseases and develop novel gene therapies. Our experiments have some limitations. We have not conducted in-depth research on microRNAs that are clearly differentially expressed, so we need to perform further experiments to confirm that and identify their functions. Moreover, whether the exosomes secreted by fibroblasts can be transported into melanocytes still needs further verification.
Author disclosure statement
The authors declare that no conflicting financial interests exist.
Declaration of patient consent
Patient’s consent not required as there are no patients in this study.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Glossary of terms
CDK2: Cyclin-dependent kinase 2, DCT: dopachrome tautomerase, DKK1: Dickkopf-1, DMEM: Dulbecco's modified eagle medium, DPBS: Dulbecco's phosphate buffered saline, ERK: extracellular signal-regulated kinase, EVs: Extracellular vesicles, FGF7: Fibroblast growth factor 7, GM-CSF: Granulocyte-macrophage Colony Stimulating Factor, GO analysis: Gene ontology analysis, GSK-3b: Glycogen synthase kinase-3b, KEGG pathway analysis: Kyoto Encyclopedia of Genes and Genomes pathway analysis, KGF: Keratinocyte growth factor, KGFR: KGF receptor, MAPK signaling pathway: Mitogen-activated protein kinase signaling pathway, MATP: Myoactive tetradeca peptide, MITF: Microphthalmia-associated transcription factor, MVBs: Multivesicular bodies, PBS: Phosphate buffered saline, piRNAs: Piwi-interacting RNA, PTN: Pleiotrophin, rRNAs: Ribosomal RNA, scRNAs: small cytoplasmic RNA, snRNAs: small nucleolar RNA, SOX5/6: SRY-box transcription factor 5/6, tRNAs: Transfer RNA, TAK1: Transforming growth factor-β activated kinase 1, TYR: Tyrosinase, TYRP1: tyrosinase-related protein 1, TPM: transcript per million
References
- CD109 is a component of exosome secreted from cultured cells. Biochem Biophys Res Commun. 2016;469:816-22.
- [CrossRef] [PubMed] [Google Scholar]
- Charge-based precipitation of extracellular vesicles. Int J Mol Med. 2016;38:1359-66.
- [CrossRef] [PubMed] [Google Scholar]
- Exosomes serve as novel modes of tick-borne flavivirus transmission from arthropod to human cells and facilitates dissemination of viral RNA and proteins to the vertebrate neuronal cells. PLoS Pathog. 2108;14:e1006764.
- [CrossRef] [PubMed] [Google Scholar]
- Exosomes are involved in mediating radiation induced bystander signaling in human keratinocyte cells. Radiat Res. 2014;181:138-45.
- [CrossRef] [PubMed] [Google Scholar]
- Exosomes released by keratinocytes modulate melanocyte pigmentation. Nat Commun. 2015;6:7506.
- [CrossRef] [PubMed] [Google Scholar]
- Identification of miR-145 as a key regulator of the pigmentary process. J Invest Dermatol. 2013;133:201-9.
- [CrossRef] [PubMed] [Google Scholar]
- miR-330-5p targets tyrosinase and induces depigmentation. J Invest Dermatol. 2014;134:2846-9.
- [CrossRef] [PubMed] [Google Scholar]
- MicroRNA-21a-5p functions on the regulation of melanogenesis by targeting sox5 in mouse skin melanocytes. Int J Mol Sci. 2016;17:E959.
- [CrossRef] [PubMed] [Google Scholar]
- MicroRNA-27a-3p inhibits melanogenesis in mouse skin melanocytes by targeting Wnt3a. Int J Mol Sci. 2015;16:10921-33.
- [CrossRef] [PubMed] [Google Scholar]
- ARP101 inhibits α-MSH-stimulated melanogenesis by regulation of autophagy in melanocytes. FEBS Lett. 2013;587:3955-60.
- [CrossRef] [PubMed] [Google Scholar]
- Human skin responses to UV radiation: Pigment in the upper epidermis protects against DNA damage in the lower epidermis and facilitates apoptosis. FASEB J. 2006;20:1486-8.
- [CrossRef] [PubMed] [Google Scholar]
- Nitric oxide produced by ultraviolet-irradiated keratinocytes stimulates melanogenesis. J Clin Invest. 1997;99:635-42.
- [CrossRef] [PubMed] [Google Scholar]
- Paracrine regulation of melanogenesis. Br J Dermatol. 2018;178:632-9.
- [CrossRef] [PubMed] [Google Scholar]
- Effects of fibroblast-derived factors on the proliferation and differentiation of human melanocytes in culture. J Dermatol Sci. 2013;71:45-57.
- [CrossRef] [PubMed] [Google Scholar]
- Skin pigmentation and pigmentary disorders: Focus on epidermal/dermal cross-talk. Ann Dermatol. 2016;28:279-89.
- [CrossRef] [PubMed] [Google Scholar]
- Precise role of dermal fibroblasts on melanocyte pigmentation. J Dermatol Sci. 2017;88:159-66.
- [CrossRef] [PubMed] [Google Scholar]
- High levels of EBV-encoded RNA 1 (EBER1) trigger interferon and inflammation-related genes in keratinocytes expressing HPV16 E6/E7. PLoS One. 2017;12:e169290.
- [CrossRef] [PubMed] [Google Scholar]
- A novel noncontact communication between human keratinocytes and T cells: Exosomes derived from keratinocytes support superantigeninduced proliferation of resting T cells. Mol Med Rep. 2017;16:7032.
- [CrossRef] [PubMed] [Google Scholar]
- Reduced MiR-675 in exosome in H19 RNA-related melanogenesis via MITF as a direct target. J Invest Dermatol. 2014;134:1075.
- [CrossRef] [PubMed] [Google Scholar]
- MicroRNA 340 is involved in UVB-induced dendrite formation through the regulation of RhoAexpression in melanocytes. Mol Cell Biol. 2014;34:3407-20.
- [CrossRef] [PubMed] [Google Scholar]
- MicroRNA-218 inhibits melanogenesis by directly suppressing microphthalmia-associated transcription factor expression. RNA Biol. 2014;11:732-41.
- [CrossRef] [PubMed] [Google Scholar]
- Regulation of pigmentation by microRNAs: MITF-dependent microRNA-211 targets TGF-β receptor 2. Pigment Cell Melanoma Res. 2105;28:217-22.
- [CrossRef] [PubMed] [Google Scholar]
- Oxidative stress-induced overexpression of miR-25: The mechanism underlying the degeneration of melanocytes in vitiligo. Cell Death Differ. 2015;23:496.
- [CrossRef] [PubMed] [Google Scholar]
- MicroRNA-155 is dysregulated in the skin of patients with vitiligo and inhibits melanogenesis-associated genes in melanocytes and keratinocytes. Acta Derm Venereol. 2016;96:742.
- [Google Scholar]
- Intrinsic and extrinsic regulation of human skin melanogenesis and pigmentation. Int J Cosmet Sci. 2018;40:328-47.
- [CrossRef] [PubMed] [Google Scholar]
- Serum levels of miRNA-21-5p in vitiligo patients and effects of miRNA-21-5p on SOX5, beta-catenin, CDK2 and MITF protein expression in normal human melanocytes. J Dermatol Sci. 2020;101:22-9.
- [CrossRef] [PubMed] [Google Scholar]
- MicroRNA-141-3p and microRNA-200a-3p regulate α-melanocyte stimulating hormone-stimulated melanogenesis by directly targeting microphthalmia-associated transcription factor. Sci Rep. 2020;10:2149.
- [CrossRef] [PubMed] [Google Scholar]
- MicroRNA 143-5p regulates alpaca melanocyte migration, proliferation and melanogenesis. Exp Dermatol. 2018;27:166-71.
- [CrossRef] [PubMed] [Google Scholar]
- miR-380-3p regulates melanogenesis by targeting SOX6 in melanocytes from alpacas (Vicugna pacos) BMC Genomics. 2019;20:962.
- [CrossRef] [PubMed] [Google Scholar]
- MicroRNA-203 regulates melanosome transport and tyrosinase expression in melanoma cells by targeting kinesin superfamily protein 5b. J Invest Dermatol. 2014;134:461-9.
- [CrossRef] [PubMed] [Google Scholar]
- SH3BP4, a novel pigmentation gene, is inversely regulated by miR-125b and MITF. Exp Mol Med. 2017;49:e367.
- [CrossRef] [PubMed] [Google Scholar]
- MicroRNA-211 regulates oxidative phosphorylation and energy metabolism in human vitiligo. J Invest Dermatol. 2017;137:1965-74.
- [CrossRef] [PubMed] [Google Scholar]
- Metabo-miR: miR-211 regulates mitochondrial energy metabolism in vitiligo. J Invest Dermatol. 2017;137:1828.
- [CrossRef] [PubMed] [Google Scholar]
- Novel inhibitory function of miR-125b in melanogenesis. Pigment Cell Melanoma Res. 2013;27:140-4.
- [CrossRef] [PubMed] [Google Scholar]






