Translate this page into:
Four novel mutations of ADAR1 in Chinese patients with dyschromatosis symmetrica hereditaria
2 Department of Dermatology, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430030; Department of Dermatology, Huangshi Central Hospital, Huangshi 435000, China
3 Department of Dermatology, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology; Department of Dermatology, The First People's Hospital of Jiangxia District, Wuhan 430030, China
4 Department of Anesthesiology, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430030, China
Correspondence Address:
Yunhua Deng
Department of Dermatology, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430030
China
| How to cite this article: Hu W, Shi X, Li H, Chen L, Wang T, Dong Y, Zhang Y, Hu M, Liu X, Zhang C, Liu D, Deng Y. Four novel mutations of ADAR1 in Chinese patients with dyschromatosis symmetrica hereditaria. Indian J Dermatol Venereol Leprol 2019;85:69-73 |
Abstract
Background: Novel mutations in adenosine deaminase acting on RNA 1 gene (ADAR1) are responsible for dyschromatosis symmetrica hereditaria (DSH). DSH patients display a mixture of hyperpigmented and hypopigmented macules on the dorsal aspects of the extremities, and freckle-like macules on the face.
Aims: To provide new evidence for further study of the etiopathogenisis of DSH.
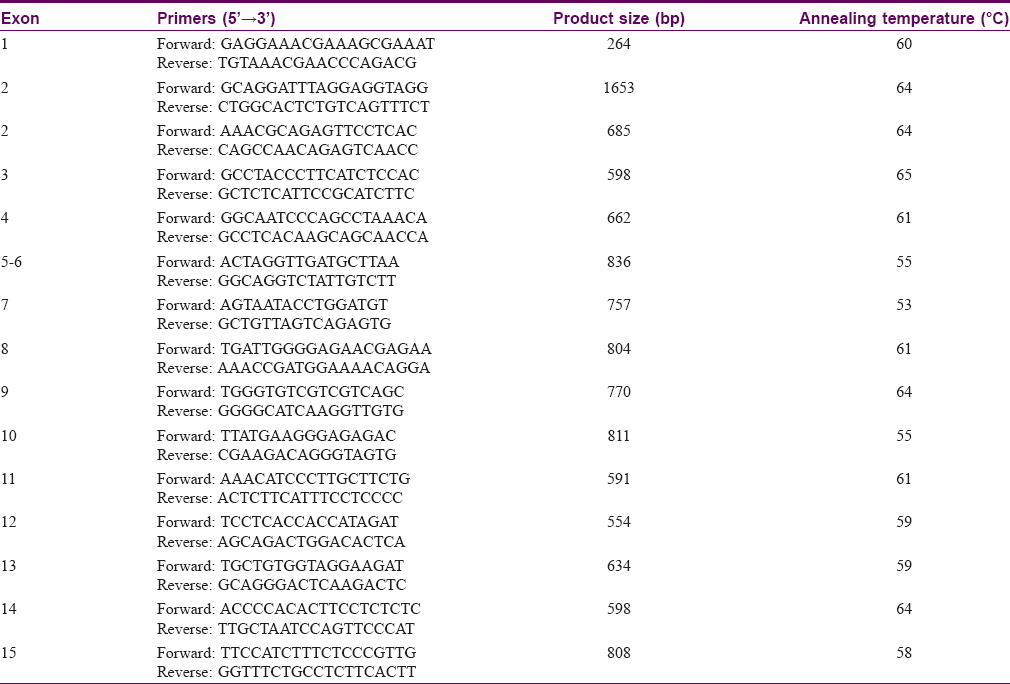
Methods: Genomic DNA was extracted and used as a template for the polymerase chain reaction (PCR) amplification of all 15 coding exons as well as intron-exon boundaries of ADAR1. The PCR products were sequenced directly.
Results: We identified eight mutations of ADAR1 in four Chinese pedigrees and four individual patients, which were c.2722G>T, p.(Asp908Tyr), c.1657delA, p.(Ser553fs), c.2563_2564delCT, p.(Leu855fs), c.526T>G, p.(Leu176Val) as well as four previously reported mutations c. 3363_3364insT, p.(Lys1122fs), c. 2865_2866delGT, p.(Val955fs), c.1630C>T, p.(Arg544X), and c.2894C>T, p.(Pro965Leu). In silico analysis predicted that all the mutations reported were pathogenic.
Limitations: We did not study how ADAR1 played its role in DSH. So, the exact pathogenic mechanism of ADAR1 in DSH patients wasn't clarified in this study.
Conclusion: We found four novel ADAR1 mutations in this study. Our results enlarge the database on ADAR1 mutations associated with DSH.
Introduction
Dyschromatosis symmetrica hereditaria (DSH; MIM # 127400) is an autosomal dominant inherited pigmentary genodermatosis characterized by intermingled hyperpigmented and hypopigmented macules on the dorsal aspect of the distal extremities, and freckle-like macules on the face. The lesions usually appear in infancy or early childhood, commonly stop spreading before adolescence, and last for life.[1] The gene responsible for DSH was identified in 2003 as adenosine deaminase acting on RNA1 (ADAR1).[2]
Methods
In this study, we studied four pedigrees of patients with DSH, four individual DSH patients whose family members refused to participate in our study or had died, and 100 unrelated normal individuals [Figure - 1]. All affected individuals had typical hyperpigmented and hypopigmented macules on their dorsal aspects of their hands and feet. The clinical features of the four probands of DSH pedigrees and four individual DSH patients are shown in [Figure - 2]. Phenotypes of all individuals were confirmed by experienced dermatologists on the basis of clinical features and family histories of DSH patients. The study was approved by the Ethics Committee of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology and written informed consent was obtained from all subjects in the study.
 |
| Figure 1: Pedigrees of the family with dyschromatosis symmetrica hereditaria and genetic findings. (a) c.2722 G>T (arrow) in exon 9 in family 1. (b) c.2865_2866delGT (arrow) in exon 10 in family 2. (e) c.1657delA (arrow) in exon 3 in family 3. (f) c.3363_3364insT (arrow) in exon 14 in family 4. (i) c.2563_2564delCT (arrow) in exon 8 of ADAR1 in individual patient 1. (j) c.526T>G (arrow) in exon 2 of ADAR1 in individual patient 2. (m) c.1630C>T (arrow) in exon 3 of ADAR1 in individual patient 3. (n) c.2894C>T (arrow) in exon 11 of ADAR1 in individual patient 4. (c), (d), (g), (h), (k), (l), (o), (p) showed relative control DNA sequences of ADAR1 from control individuals, respectively |
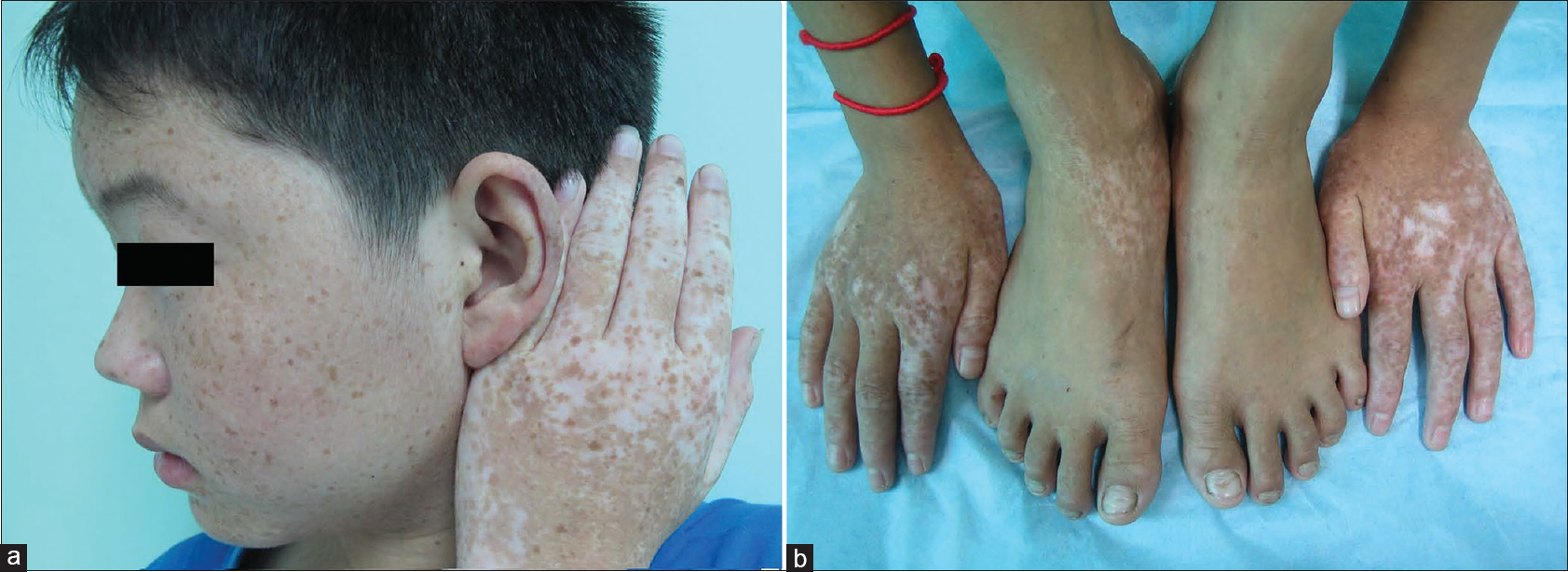 |
| Figure 2: Typical skin lesions of the patients with dyschromatosis symmetrica hereditaria. (a) Intermingled hyper-pigmented and hypo-pigmented maculeson the dorsal aspect of the hands and freckle-like macules on the face of the DSH patient. (b) Intermingled hyper-pigmented and hypo-pigmented macules onthe dorsal aspect of the DSH patient's distal extremities |
Genomic DNA was extracted from peripheral blood samples of 14 DSH patients, six clinically unafffected family members in the four pedigrees, and 100 control individuals. All 15 exons of ADAR1 and their flanking intron sequences were amplified by polymerase chain reaction (PCR) using specific primers designed by Primer Premier 5 [Table - 1]. PCR was performed as previously described.[3] After the amplification, the products were purified using a QIAquick PCR Purification Kit (Qiagen, Hilden, Germany) and sequenced using an ABI PRISM® 3730 automated sequencer (Applied Biosystems, Foster City, CA, USA). Sequence comparisons and analyses were performed using the Basic BLAST program. Mutations were identified by comparison with the reported cDNA reference sequences for ADAR1 (GenBank accession number: NM_001111.4). Then all ADAR1 mutations previously reported were collected according to PubMed and Human Gene Mutation Database (HGMD). Eight different heterozygous mutations of ADAR1 were identified in this study [Table - 2].

Results
In family 1, a missense mutation, c.2722 G>T, p.(Asp908Tyr) in exon 9 of ADAR1 was identified in the proband, his mother and his daughter, but not in unaffected members of his family and 100 control individuals [Figure - 1]a and [Figure - 1]c. The same running procedure was performed and a deletion mutation, c.2865_2866delGT, p.(Val955fs) in exon 10 of ADAR1 was found in family 2 [Figure - 1]b and [Figure - 1]d. Family 3 had a deletion mutation c.1657delA, p.(Ser553fs) in exon 3 of ADAR1 [Figure - 1]e and [Figure - 1]g. Family 4 harbored an insertion mutation c.3363_3364insT, p.(Lys1122fs) in exon 14 of ADAR1 [Figure - 1]f and [Figure - 1]h. Individual patient 1 had a deletion mutation c.2563_2564delCT, p.(Leu855fs) in exon 8 of ADAR1 [Figure - 1]i and [Figure - 1]k. Individual patient 2 carried a missense mutation c.526T>G, p.(Leu176Val) in exon 2 of ADAR1 [Figure - 1]j and [Figure - 1]l. Individual patient 3 carried a nonsense mutation c.1630C>T, p.(Arg544X) in exon 3 of ADAR1 [Figure - 1]m and [Figure - 1]o. Individual patient 4 had a missense mutation c.2894C>T, p.(Pro965Leu) in exon 11 of ADAR1 [Figure - 1]n and [Figure - 1]p. Four mutations c.2865_2866delGT, p.(Val955fs), c.3363_3364insT, p.(Lys1122fs), c.1630C>T, p.(Arg544X) and c.2894C>T, p.(Pro965Leu) of ADAR1 have been reported previously.[4],[5],[6],[7] All eight mutations were proved to be pathogenic by Mutation Taster, Provean, and SIFT.
Discussion
ADAR1 consists of 1,226 amino acid residues and contains at least six functional domains: Two copies of a Z-DNA-binding domain (Zα and Zβ), three copies of the double stranded ribonucleic acid (dsRNA)-binding domain (RI, RII, and RIII) and a transfer RNA (tRNA)-specific and dsRNA adenosine deaminase domain (ADEAMc).[8] It has been discovered to possess different functions in RNA editing and RNA interference (RNAi) by the formation of either ADAR1/ADAR1 homodimer or Dicer/ADAR1 heterodimer complexes, respectively.[9]
In this study, we identified four novel ADAR1 mutations in four pedigrees and four individual patients with DSH. To date, 196 different ADAR1 mutations have been found in patients with DSH around the world including our data. According to the functional domain of ADAR1, three (1.5%) mutations among them are located in the Zα domain, 5 (2.6%) in the Zβ domain, 13 (6.6%) in the RI domain, four (2.0%) in the RII domain, seven (3.6%) in the RIII domain and 103 (52.6%) within the ADEAMc domain. These results clearly suggest that the ADEAMc domain might be a hot spot for ADAR1 mutations associated with DSH. This is in accord with the finding reported by Li.[10] The ADEAMc domain of ADAR1 has been verified to be highly conserved in different species. Any mutation in the domain would result in dysfunction of adenosine deaminase, but the exact mechanisms involved are still mysterious.
In summary, our findings expanded the spectrum of ADAR1 mutations associated with DSH and could be a great help for future clinical genetic counseling.
There were also some limitations in our research. We did not study the exact pathogenic mechanism of ADAR1 in DSH patients. So, further research is needed to clarify the function of ADAR1.
Acknowledgments
We would like to thank all the subjects that participated in this study.
Declaration of patient consent
The authors certify that they have obtained all appropriate patient consent forms. In the form, the patient has given his consent for his images and other clinical information to be reported in the journal. The patient understand that name and initials will not be published and due efforts will be made to conceal identity, but anonymity cannot be guaranteed.
Financial support and sponsorship
This work was supported financially by Grants from National Natural Science Foundation of China (No. 81371728, No. 81472864).
Conflicts of interest
There are no conflicts of interest.
| 1. |
Hayashi M, Suzuki T. Dyschromatosis symmetrica hereditaria. J Dermatol 2013;40:336-43.
[Google Scholar]
|
| 2. |
Miyamura Y, Suzuki T, Kono M, Inagaki K, Ito S, Suzuki N, et al. Mutations of the RNA-specific adenosine deaminase gene (DSRAD) are involved in dyschromatosis symmetrica hereditaria. Am J Hum Genet 2003;73:693-9.
[Google Scholar]
|
| 3. |
Liu Q, Wang Z, Wu Y, Cao L, Tang Q, Xing X, et al. Five novel mutations in the ADAR1 gene associated with dyschromatosis symmetrica hereditaria. BMC Med Genet 2014;15:69.
[Google Scholar]
|
| 4. |
Okamura K, Abe Y, Fukai K, Tsuruta D, Suga Y, Nakamura M, et al. Mutation analyses of patients with dyschromatosis symmetrica hereditaria: Ten novel mutations of the ADAR1 gene. J Dermatol Sci 2015;79:88-90.
[Google Scholar]
|
| 5. |
Wang XP, Wang WJ, Wang JM, Liu Y, Xiao SX. Four novel and two recurrent mutations of the ADAR1 gene in Chinese patients with dyschromatosis symmetrica hereditaria. J Dermatol Sci 2010;58:217-8.
[Google Scholar]
|
| 6. |
Suzuki N, Suzuki T, Inagaki K, Ito S, Kono M, Fukai K, et al. Mutation analysis of the ADAR1 gene in dyschromatosis symmetrica hereditaria and genetic differentiation from both dyschromatosis universalis hereditaria and acropigmentatio reticularis. J Invest Dermatol 2005;124:1186-92.
[Google Scholar]
|
| 7. |
Yuan C, Liu H, Fu X, Yu Y, Yu G, Bao F, et al. Two novel mutations of the ADAR1 gene in Chinese patients with dyschromatosis symmetrica hereditaria. Indian J Dermatol Venereol Leprol 2012;78:746-8.
[Google Scholar]
|
| 8. |
George CX, Gan Z, Liu Y, Samuel CE. Adenosine deaminases acting on RNA, RNA editing, and interferon action. J Interferon Cytokine Res 2011;31:99-117.
[Google Scholar]
|
| 9. |
Ota H, Sakurai M, Gupta R, Valente L, Wulff BE, Ariyoshi K, et al. ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell 2013;153:575-89.
[Google Scholar]
|
| 10. |
Li M, Yang L, Li C, Jin C, Lai M, Zhang G, et al. Mutational spectrum of the ADAR1 gene in dyschromatosis symmetrica hereditaria. Arch Dermatol Res 2010;302:469-76.
[Google Scholar]
|
Fulltext Views
3,213
PDF downloads
2,515





