Translate this page into:
Ligand-binding prediction for ErbB2, a key molecule in the pathogenesis of leprosy
Correspondence Address:
Viroj Wiwanitkit
Department of Clinical Laboratory Medicine, Faculty of Medicine, Chulalongkorn University, Bangkok- 10330
Thailand
| How to cite this article: Wiwanitkit V. Ligand-binding prediction for ErbB2, a key molecule in the pathogenesis of leprosy. Indian J Dermatol Venereol Leprol 2008;74:32-34 |
Abstract
Background and Aims: Mycobacterium leprae is an obligate intracellular pathogen. Ligand-binding is an important factor in the success of chemoprevention and chemotherapy. A new drug that can inhibit M. leprae binding to and activation of, ErbB2 and Erk1/2 in primary Schwann cells is the new therapeutic option. However, the ligand-binding pattern of ErbB2 has never been clarified. Methods: In this work, the author performed a ligand-binding prediction for ErbB2 using a new bioinformatics tool. Results: According to this study, nine strong possible ligands can be identified. Conclusion: These sites can be useful for further drug-development studies.

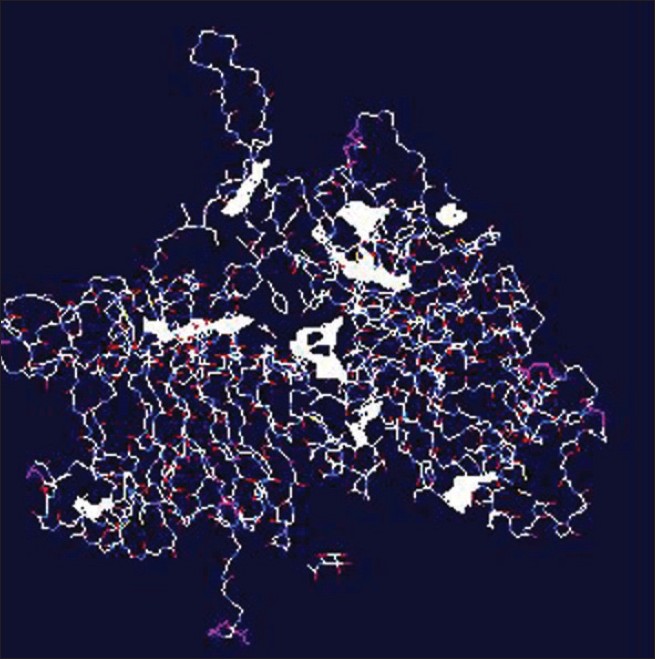 |
| Figure 1: The predicted ligands within ErbB2 *The area in sheath plate is the identifi ed area (ligand) |
 |
| Figure 1: The predicted ligands within ErbB2 *The area in sheath plate is the identifi ed area (ligand) |
Introduction
Cell adhesion plays a pivotal role in diverse biological processes that occur in the dynamic setting of an infection. [1] Although complex, cell adhesion in the vasculature can be exploited to direct drug carriers to targeted cells and tissues. Ligand-binding is an important factor in the pathogenesis of many infections. [2] Ligand-binding analysis becomes an important novel tool for new drug discovery. The analysis can be useful for identification of the point of interaction and can be a clue for further molecular docking study.
M. leprae, the causative agent of leprosy, is an obligate intracellular bacterial pathogen. Basically, M. leprae-induced demyelination is a result of direct pathogen ligation to, and activation of, ErbB2 receptor tyrosine kinase signaling through a complex process. [3],[4] Herceptin ® (trastuzumab), a therapeutic humanized ErbB2-specific antibody that can inhibit M. leprae binding to and activation of, ErbB2 and Erk1/2 in primary Schwann cells, might turn out to be a new therapeutic option. [4],[5] Ligand-binding is an important factor in the success of chemoprevention and chemotherapy of leprosy. [6] However, the ligand-binding pattern of ErbB2 has never been clarified. In this work, the author performed a ligand-binding prediction for ErbB2 using a new bioinformatics tool.
Methods
Getting the sequence
The database Pubmed was used for data mining of the molecular structure for ErbB2 receptor.
Prediction for the ligand-binding
The ligand-binding for ErbB2 receptor was performed using a novel bioinformatics tool, namely, Q-SiteFinder. [7] It is a new method of ligand-binding site prediction. [7] It uses the interaction energy between the protein and a simple van der Waals probe to locate energetically favorable binding sites. [7] Energetically favorable probe sites are clustered according to their spatial proximity and clusters are then ranked according to the sum of interaction energies for sites within each cluster. [7] The system works by scanning a probe radius of 1.6 Å along all gridlines of a grid resolution 0.9 Å surrounding the protein. Grid points are defined to be part of a site when the probe is within the range of protein atoms followed by free space, followed by protein atoms. [7] If ligands are allowed to remain on the protein, they may interfere with binding-site prediction. The proteins are initially scanned for ligands and then, potential ligands are identified. [7] The method can be applied in structural genomics studies where protein-binding sites remain uncharacterized since the 86% success rate for unbound proteins appears to be only slightly lower than that of ligand-bound proteins. [7]
Visualization of the ligand
The result was further processed to build a format of three-dimensional (3D) molecular structure by Swiss-Pdb Viewer [8] (GlaxoSmithKline ® and Swiss Institute of Bioinformatics ® ). Briefly, Swiss-Pdb Viewer is used to generate 3D molecular structure, side by side stereo and above/below stereo. [8] Based on the two distant stereo modes, recomposition of data file to 3D structure can be done.
Results
The molecular structure of the ErbB2 receptor (2A91) was derived and used as template for further prediction for ligand-binding sites. According to the analysis, nine ligands can be identified. The results from the analysis are shown in [Table - 1] and [Figure - 1].
Discussion
Pathogens have co-evolved with their hosts and acquired the ability to intercept, disrupt, mimic and usurp numerous signaling pathways of those hosts. [1] The study of host-pathogen interactions not only teaches us about the intricate biology of these parasitic invaders but also provides interesting insights into basic cellular processes both at the individual cell level and more globally throughout the organism. [1] There are many steps in pathogenesis of each infection. These include access to an infecting surface, multiplication on that surface, colonization, invasion, multiplication inside the new host and resistance to innate and adaptive immune mechanisms. [2] Many such steps require a specific ligand-receptor interaction. [2]
M. leprae can infect a variety of cells in vivo, including epithelial cells, muscle cells and Schwann cells, in addition to macrophages. [6] The area (ligand) receptor interactions are important in the attachment and ingestion of M. leprae by these nonmacrophage cells. [6] Recent work by Tapinos et al. [4] shows that a direct mechanism of demyelination induced by M. leprae depends on the binding of the pathogen to the receptor tyrosine kinase ErbB2 on Schwann cells and the resulting activation of the Ras-Raf-MEK-ERK pathway. [5] Blockade of ErbB2 activity by the new molecule that acts on the ligand-receptor interaction might be an effective therapeutic tool. For example, a new molecule PKI-166, a specific inhibitor of EGF-R phosphorylation, dual ErbB1-ErbB2 kinase, can effectively abrogate M. leprae-induced myelin damage in both in vitro and in vivo models. [3],[4],[5]
Analysis of the ligand-binding within ErbB2 molecule can be useful for new preventive and therapeutic drug development for leprosy. With the increasing amount of data provided by both high-throughput sequencing and structural genomics studies, there is a growing need for tools to augment functional predictions for protein sequences. Broad descriptions of function can be provided by establishing the presence of protein domains associated with a particular function. Here, the author used a bioinformatics tool for analysis of the ligand-binding pattern within the ErbB2 molecule.
According to this study, nine strong possible ligands can be identified. Indeed, there are only a few reports on the ligand-binding of ErbB2 in leprosy. [3],[4] Several new ligands are detected in this work. These sites can be useful for further drug development studies. Prediction of the roles and vulnerable binding descriptions of these sites are planned in the future research.
| 1. |
Munter S, Way M, Frischknecht F. Signaling during pathogen infection. Sci STKE 2006;2006:re5.
[Google Scholar]
|
| 2. |
Sansonetti P. How to define the species barrier to pathogen transmission? Bull Acad Natl Med 2006;190:611-22.
[Google Scholar]
|
| 3. |
Franklin RJ, Zhao C. Tyrosine kinases: Maiming myelin in leprosy. Nat Med 2006;12:889-90.
[Google Scholar]
|
| 4. |
Tapinos N, Ohnishi M, Rambukkana A. ErbB2 receptor tyrosine kinase signaling mediates early demyelination induced by leprosy bacilli. Nat Med 2006;12:961-6.
[Google Scholar]
|
| 5. |
Noon LA, Lloyd AC. Treating leprosy: An Erb-al remedy? Trends Pharmacol Sci 2007;28:103-5.
[Google Scholar]
|
| 6. |
Schorey JS, Li Q, McCourt DW, Bong-Mastek M, Clark-Curtiss JE, Ratliff TL, et al. A Mycobacterium leprae gene encoding a fibronectin binding protein is used for efficient invasion of epithelial cells and Schwann cells. Infect Immun 1995;63:2652-7.
[Google Scholar]
|
| 7. |
Laurie AT, Jackson RM. Q-SiteFinder: An energy-based method for the prediction of protein-ligand binding sites. Bioinformatics 2005;21:1908-16.
[Google Scholar]
|
| 8. |
Kaplan W, Littlejohn TG. Swiss-PDB viewer (deep view). Brief Bioinform 2001;2:195-7.
[Google Scholar]
|
Fulltext Views
2,570
PDF downloads
3,692





