Translate this page into:
The role of glucose-dependent insulinotropic polypeptide 3 (G1P-3) and nucleolar phosphoprotein-1 (NPM1) in pathogenesis of psoriasis
Corresponding author: Prof. Olfat G. Shaker, Department of Medical Biochemistry and Molecular Biology, Faculty of Medicine, Cairo University, Cairo, Egypt. olfatshaker@yahoo.com
-
Received: ,
Accepted: ,
How to cite this article: Hassan M H, ElTahlawi S, Shaker OG, Magdy M. The role of glucose-dependent insulinotropic polypeptide 3 (G1P-3) and nucleolar phosphoprotein-1 (NPM1) in pathogenesis of psoriasis. Indian J Dermatol Venereol Leprol 2023;89:828-33.
Abstract
Background
Psoriasis is a multifactorial, hyperproliferative, chronic inflammatory skin disease affecting males and females equally.
Aims
To study the expression of certain non-coding RNAs, Interferon Alpha Inducible Protein 6 (IFI6), previously named Glucose-dependent Insulinotropic Polypeptide 3 (G1P-3), and nucleolar phosphoprotein (in serum and tissue), and to attempt to elucidate their role in the pathogenesis of psoriasis, which in turn might help in treatment.
Methods
Twenty patients with psoriasis and 20 healthy subjects were included in this study. Serum and skin biopsies were obtained from all participants. Molecular biology techniques were employed to estimate the expression levels of long noncoding G1P-3 and nucleolar phosphoprotein in serum and skin biopsy.
Results
Psoriasis patients had a mean age of 41.85 ± 12.29. The median serum G1P-3 level of the patients’ group (3.330) was significantly higher than that of the control group (1.085) (P ≤ 0.001). Tissue G1P-3 level of the patients’ group (6.495) was also significantly higher compared to that of controls (1.040) (P ≤ 0.001). Similarly, for nucleolar phosphoprotein, the median serum level of patients’ group (2.030) was significantly higher than that of controls (1.040) (P ≤ 0.001) and median tissue level (5.425) was also significantly higher than that of controls (1.040) (P ≤ 0.001).
Limitations
In this study, only outpatients were included and follow-up was not well-handled. For future work, follow-up can be considered.
Conclusion
Long non-coding G1P-3 as well as nucleolar phosphoprotein may be considered as genetic markers for psoriasis susceptibility. In future, these might provide a novel direction for advances in psoriasis treatment.
Keywords
Psoriasis
pathogenesis
genetics
Plain Language Summary
Psoriasis is a hot topic of concern in many countries. It is a chronic inflammatory skin disease affecting males and females equally. This is a clinical and biochemical study that explores the expression of some non-coding RNAs (including long noncoding RNAs) namely, glucose-dependent insulinotropic polypeptide 3 (G1P-3) and nucleolar phosphoprotein in both serum and tissue in order to determine their role in the pathogenesis of psoriasis, which in turn might help in treatment. Laboratory and pathological investigations were undertaken. Results revealed that psoriatic patients had high expression of both biomarkers in serum and tissue as compared to the healthy control group.
Introduction
Psoriasis is reported to be one of the most widely prevalent chronic systemic inflammatory diseases presenting with both cutaneous and systemic manifestations. More than 2% of the general population all over the world is affected by the disease.1,2 The occurrence and progression of psoriasis are affected by multiple factors and inheritance is polygenic. Non-coding RNAs play a vital role in individual susceptibility to psoriasis. It has been proven that there is dysregulation of long-non-coding RNAs in the peripheral blood and even in the skin lesions of psoriatic patients.3,4
Stress is a common trigger for psoriasis flares. PRINS is a non-coding RNA that regulates G1P3, a gene with anti-apoptotic effects in keratinocytes. The approved name for PRINS is psoriasis associated non-protein coding RNA induced by stress. Deregulation of the PRINS non-coding RNA may contribute to psoriasis.5 In silico structural and homology studies suggested that PRINS may function as a noncoding RNA. It harbours two alu elements, it is transcribed by RNA polymerase II, and it is expressed at different levels in various human tissues.6
PRINS is reported to be overexpressed in both lesional and non-lesional psoriatic epidermis that is induced by stress.5,6 PRINS was expressed higher in the uninvolved epidermis of psoriatic patients compared with both psoriatic lesional and healthy epidermis, suggesting a role for PRINS in psoriasis susceptibility.6
Nucleolar phosphoprotein has a potential role as a regulator of cell proliferation. It is considered a physical interactor with PRINS. Immuno-histochemical experiments have shown a high expression of nucleolar phosphoprotein in the cells of the basal layers of lesional psoriatic skin as opposed to healthy and non-lesional psoriatic skin.7 In UVB-treated cells, nucleolar phosphoprotein is transported into the nucleus. The elimination of the PRINS gene causes nucleolar phosphoprotein to remain in the nucleoli of keratinocytes irradiated with UV-B.8 Nucleolar phosphoprotein also has a potential role in chronic inflammatory skin diseases such as atopic dermatitis and malignant cutaneous melanoma.9,10
In the present study, the gene Interferon Alpha Inducible Protein 6 (IFI6) or G1P-3 as well as nucleolar phosphoprotein were assessed in the serum and tissue of Egyptian psoriatic patients and compared with that of healthy controls, in an attempt to elucidate their role in the pathogenesis of psoriasis, which in turn might help in developing newer methods of treatment.
Methods
The current study was conducted on 20 patients with psoriasis and 20 apparently healthy subjects who served as controls with matching age & sex, during the period from January 2020 to January 2021. Patients were recruited from the dermatology department, faculty of medicine, Fayoum University. Ethical approval was taken from faculty of medicine, Fayoum University(no. D110). The duration of disease was variable, ranging from 8 to 360 months with a mean ± standard deviation of 122.55 ± 106.91 months.
Inclusion criteria
All included patients had plaque psoriasis.
Exclusion criteria
Patients with any other chronic inflammatory or autoimmune diseases, patients with metabolic diseases, any type of cancer, pregnant and lactating females were excluded. Patients were all naïve and had not been on any treatment for psoriasis.
Written consent was obtained from each participant before enrolment in the study. Detailed history of psoriasis was taken including details of onset, course and duration. Clinical assessment of the type and extent of psoriasis as well as Psoriasis Area and Severity Index (PASI) were undertaken.11
From each patient and control, serum and a 4 mm skin punch biopsy were obtained.
Molecular biology techniques were employed for estimation of long noncoding G1P-3 and nucleolar phosphoprotein expression levels in serum and skin biopsy
miRNeasy mini kit was used for purification of long noncoding RNA (Qiagen, Valencia, CA, USA). RNA quantitation and purity was assessed by NanoDrop® (ND)—1000 spectrophotometer. RNA was reverse transcribed into cDNA using the miScript II RT kit (Qiagen, Valencia, CA, USA) as follows:
Hiflex buffer, Nucleics mix, miScrippt Reverse transcriptase mix and RNA (60 ug).
Gentle mixing was done by centrifugation. Incubation was done for 60 minutes at 37ºC using conventional PCR. Incubation for 5 min at 95ºC was done to inactivate Reverse Transcriptase using conventional PCR.
Detection of both biomarkers
Fold change for both long noncoding RNAs were determined using readymade primers and customized GAPDH as internal control housekeeping gene for all cases and healthy controls. GAPDH F: 5′-CCCTTCATTGACCTCAACTA-3′, GAPDH-R: 5′-TGGAAGATGGTGATGGGATT-3′. Real-time PCR was performed using Rotor Gene Q System (Qiagen). Fold change was calculated using 2−ΔΔCt relative to control samples.
Statistical analysis of data
The analysis of the data was performed by using SPSS 17 on windows 8.1. For the descriptive statistics, mean ± standard deviation was used for parametric numerical data. For non-parametric numerical data, median was used. t-test was used to measure the significance of differences in parametric data obtained from the two groups, while U test (Mann-Whitney) was used similarly for to measure the non-parametric variables. Chi-square test was used to show the relationship between qualitative variables. For continuous variables, Pearson-r correlation was used to test the association between groups (serum and tissue samples from controls and psoriatic patients respectively) with a two-tailed test of significance. Sensitivity and specificity of measuring G1P-3 and nucleolar phosphoprotein were analysed using a Receiver Operating Characteristic (ROC) Curve. For all statistical analyses, P-values <0.05 were considered significant
Results
Demographic and clinical characteristics of study population
Serum and tissue samples from 20 healthy controls (including 15 males and 5 females, mean age 41.85 ± 12.29 years), and 20 patients with psoriasis (including 14 males and 6 females, mean age 41.30 ± 12.54 years) were enrolled in this study. No statistical significance was observed with respect to age or sex of subjects in the two groups with P-values of 0.88 and 0.72, respectively. Among the studied subjects, (20%) had a family history of psoriasis, but there was no statistically significant difference between patients and controls (P-value = 0.26).
Biomarker levels in psoriasis and control groups
Nucleolar phosphoprotein and G1P-3 gene expression were examined in serum and tissue of the psoriasis group as well as the healthy control group. Both expressions were higher in the psoriasis group and the differences were all highly statistically (P-value <0.001) [Table 1].
| Biomarkers | Psoriasis | Control | Mann-Whitney U Statistic | P-value |
|---|---|---|---|---|
| (Median) | ||||
| NPM (serum) | 2.030 | 1.040 | 31.500 | <0.001a |
| G1P-3 (serum) | 3.330 | 1.085 | 38.000 | <0.001a |
| NPM (tissue) | 5.425 | 1.040 | 610.000 | <0.001a |
| G1P-3 (tissue) | 6.495 | 1.040 | 610.000 | <0.001a |
P values in bold are statistically significant (P < 0.05). aOne-way, NPM: Nucleolar phosphoprotein, G1P-3: Glucose-1 phosphate-3
Relationship between nucleolar phosphoprotein, G1P-3 and different characteristics in the psoriasis group
With regards to the onset of disease, we categorized the psoriasis patients into two different groups, i.e., those with gradual onset and those with sudden onset. For serum biomarker levels, results revealed that there was no statistically significant difference between the gradual onset and sudden onset groups (P-value = 0.795 and 0.754, for nucleolar phosphoprotein and G1P-3 respectively). The same was the case with tissue levels of both biomarkers (P-values 0.146 and 0.879, respectively) [Table 2].
| Clinical data | NPM (serum) | NPM (tissue) | G1P-3 (serum) | G1P-3 (tissue) |
|---|---|---|---|---|
| Disease onset | ||||
| Gradual | 2.87 ± 0.34 | 5.87 ± 2.33 | 3.63 ± 1.90 | 6.54 ± 2.16 |
| Sudden | 2.66 ± 0.80 | 3.92 ± 1.02 | 3.30 ± 1.41 | 6.72 ± 1.45 |
| Disease course | ||||
| Remissions & exacerbations | 2.83 ± 0.42 | 5.29 ± 2.49 | 3.07 ± 1.92 | 6.25 ± 1.91 |
| progressive | 2.82 ± 0.45 | 5.84 ± 2.24 | 3.88 ± 1.52 | 7.18 ± 2.24 |
| Family history | ||||
| Positive | 2.91 ± 0.79 | 3.29 ± 0.69 | 6.58 ± 2.54 | 6.00 ± 1.37 |
| Negative | 2.44 ± 0.25 | 3.37 ± 0.99 | 5.20 ± 2.04 | 6.72 ± 2.12 |
NPM: Nucleolar phosphoprotein, G1P-3: Glucose-1 phosphate-3
When patients were categorised by disease course (remitting - relapsing versus progressive), there was also no statistical significance between serum levels of nucleolar phosphoprotein (P-value = 0.992) or G1P-3 (P-value = 0.175). The same was true for tissue levels of both biomarkers [Table 2].
There was also no significant link between family history and levels of NPM1 or G1P3 biomarkers in serum or tissue [Table 2].
There was no statistical correlation between the age of patients and biomarker levels in serum or tissue with (r, P-values) of (-0.24, 0.30), (0.053, 0.82), (-0.20, 0.37) and (0.11, 0.63), respectively.
Relationship of serum and tissue NPM1 and G1P-3 levels with disease severity in psoriasis patients
Based on the scores, psoriasis patients were classified into four groups: mild (10%), moderate (70%), severe (15%) and very severe (5%). Results revealed that when the PASI score increases both biomarkers levels in serum and tissue as well overexpress. The mean levels of nucleolar phosphoprotein in serum were 1.27 ± 0.04, 2.93 ± 0.39, 11.93 ± 0.29, 18.58 ± 0.01 for mild, moderate, severe and very severe, respectively. For tissue levels of nucleolar phosphoprotein, 2.43 ± 0.65, 5.7 ± 0.66, 19.62 ± 2.23, 26.53 ± 0.01 for all groups, respectively [Figure 1]. Results revealed a statistically significant difference between mild and moderate groups (P-value = 0.001), mild and severe groups (P-value <0.001) as well as mild and very severe groups (P-value=0.003). Likewise, there were also statistically significant differences in serum nucleolar phosphoprotein levels between moderate and severe groups and also between moderate and very severe groups with P-value <0.001.
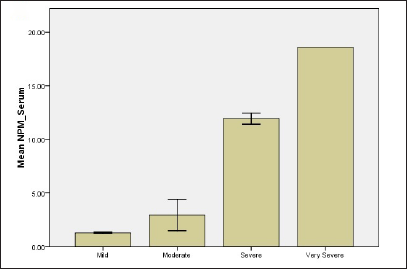
- Relationship of NPM and G1P-3 levels of psoriatic patients with regard to disease severity (a) NPM level in serum
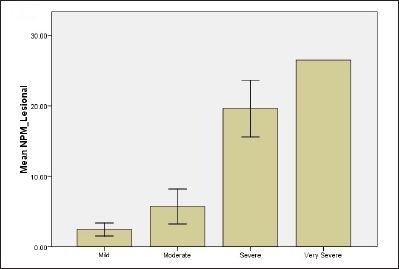
- Relationship of NPM and G1P-3 levels of psoriatic patients with regard to disease severity (b) NPM level in lesional skin
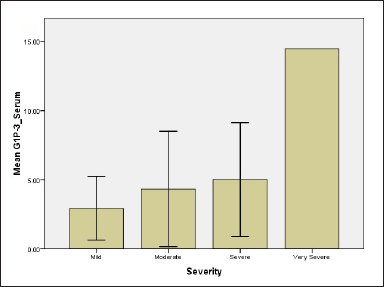
- Relationship of NPM and G1P-3 levels of psoriatic patients with regard to disease severity (c) G1P-3 level in serum
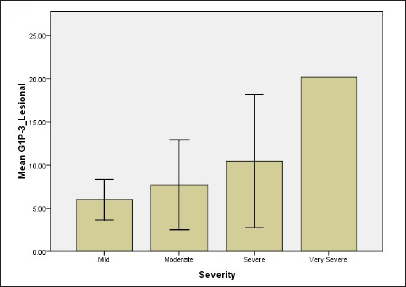
- Relationship of NPM and G1P-3 levels of psoriatic patients with regard to disease severity (d) G1P-3 level in lesional skin
While regarding G1P-3 levels in serum and tissue 2.94 ± 1.61, 4.33 ± 1.11, 5.0 ± 2.3, 14.4 ± 0.01 for serum for all groups and 5.98 ± 1.66, 7.7 ± 1.39, 10.44 ± 4.4, 20.18 ± 0.001 for tissue, respectively [Figure 1]. Results showed significant differences between mild and moderate groups (P-value = 0.05), mild and severe groups (P-value = 0.04), mild and very severe groups (P-value <0.001) as well as moderate and very severe groups (P-value = 0.036). The difference in tissue levels of G1P-3 were significant between mild and very severe groups (P-value -0.012) and also between moderate and very severe groups (P-value = 0.038).
Correlations between nucleolar phosphoprotein and G1P-3 in serum and tissue
A positive correlation was found between nucleolar phosphoprotein expression levels in serum and tissue (r = 0.944, P-value <0.001). Similarly, there was also a positive correlation between G1P-3 levels in serum and in tissue (r=0.958, P-value <0.001). In addition, there was a positive correlation between nucleolar phosphoprotein and G1P-3 levels in tissue (r = 0.556, P-value = 0.011).
ROC curve analysis for NPM1 and G1P-3 (serum and tissue)
Receiver Operating Characteristics (ROC) curve analysis was undertaken for nucleolar phosphoprotein and G1P-3 serum and tissue levels. It was found that each one of these four markers had excellent abilities to discriminate between psoriasis patients and controls. For serum nucleolar phosphoprotein level, the area under the curve (AUC) was 0.98 with sensitivity 93.2% and specificity 99% at a cut-off value of 2.43-fold change. For tissue nucleolar phosphoprotein level, the AUC was 0.99 with sensitivity 98% and specificity 98.2% at a cut-off value of 5.67-fold change. For serum G1P-3 level, the AUC was 0.97 with sensitivity 95% and specificity 98.5% at a cut-off value of 3.48-fold change. For tissue GIP-3 level, the AUC was of 0.99 with sensitivity 98.2% and specificity 99% at a cut-off value of 6.56-fold change [Figure 2].
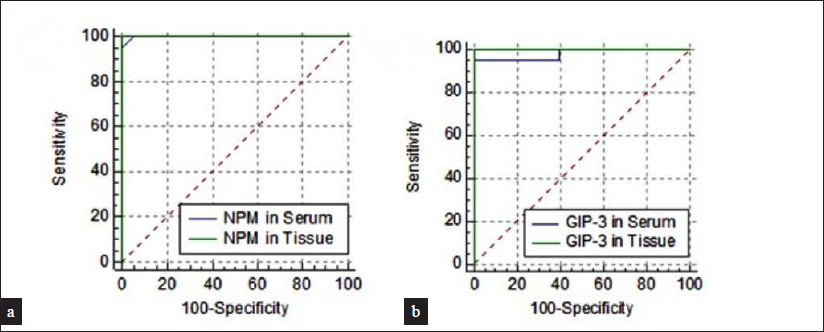
- ROC curve for (a) NPM levels in serum and tissue, (b) GIP-3 in serum and tissue among psoriasis patients
Discussion
PRINS is the main gene that regulates both G1P-3 and nucleolar phosphoprotein. G1P3 has a potential role in the pathogenesis of psoriasis.7 The deregulation of the PRINS non-coding RNA contributes to psoriasis and results in decreased sensitivity to spontaneous keratinocyte apoptosis via the regulation of G1P-3. PRINS regulates it, a gene with anti-apoptotic effects, in keratinocytes. Also, nucleolar phosphoprotein is a potential physical interactor with PRINS.7
In this study, we aimed to investigate the expression level of nucleolar phosphoprotein and G1P-3 in serum and lesional skin across our groups. Szegedi et al. demonstrated that PRINS changes cell morphology as well as gene expression profile.5 G1P-3 has a vital role in the pathogenesis of psoriasis, it regulates the anti-apoptotic gene G1P-3 in keratinocytes.5,12 PRINS regulates G1P-3, it was previously found that G1P-3 was upregulated in hyperproliferative lesions and also upregulated in non-lesional epidermis compared to healthy controls.10,12
We found that G1P-3 was over-expressed in the tissue of psoriatic patients. Our results suggested a vital role for G1P-3 in psoriasis susceptibility and in cellular stress response. Based on the experimental results in this study, we suspect that G1P-3 might act as a regulatory RNA that modifies the expression of protein-coding genes. This hypothesis is supported by other studies which showed that up-regulation of G1P-3 promotes proliferation and suppresses apoptosis.12-14 Various prior studies have illustrated the importance of PRINS non-coding RNA gene in psoriatic patients.14-16
We next analysed the expression of nucleolar phosphoprotein in the tissue of psoratic patients. It was found that the expression of nucleolar phosphoprotein was remarkably increased in psoriatic epidermis.
Other studies agree with our results17,18 and state that nucleolar phosphoprotein is overexpressed in psoriatic involved epidermis. Previous observations reveal that it has a vital role in normal keratinocyte proliferation and that its over-expression occurs in certain hyper-proliferative and malignant skin diseases, which is coherent with our findings.7
Previously, transforming growth factor beta (TGF-β1) or vascular endothelial growth factor A (VEGF-A) have been proposed as useful therapeutic targets for atopic dermatitis. Several reports have mentioned the important role and association of nucleolar phosphoprotein in the TGF-β1 pathway for skin diseases. One group of authors concluded that nucleolar phosphoprotein provides a novel insight into TGF-β1- as well as VEGF-A-related diseases.9 Another study found increased tissue and serum levels of TGF-β1 in psoriasis patients.2 It has also been shown that VEGF is an important factor for angiogenesis in psoriasis.19
We were unable to find any previous reports to determine the relationship between serum and tissue expression of G1P-3 & nucleolar phosphoprotein in psoriatic patients in relation to clinical data. Our results revealed that there was no statistically significant difference between G1P-3 and nucleolar phosphoprotein in serum or in tissue with regard to age, sex, family history, disease onset or even the course of the disease.
We also explored the relationship between PASI score and the biomarkers. Results revealed that there were significant differences in nucleolar phosphoprotein and G1P-3 levels, both in serum and tissue, when patients with different severities of psoriasis were compared. A number of these differences were highly statistically significant (discussed in greater detail in the results section). We generally found that the expression of all 4 biomarkers uptrended as PASI scores increased.
On further analysis, we found a positive correlation between the levels of serum and tissue nucleolar phosphoprotein levels, between serum and tissue G1P-3 levels as well as between both biomarkers in tissue.
Limitations
There is a lack of previous research studies on the role of nucleolar phosphoprotein and G1P-3 on the pathogenesis of psoriasis. This study had a limited sample size.
Conclusion
Both G1P-3 and high nucleolar phosphoprotein expression were found to be increased in psoriatic epidermis. This suggests that they are vital in the pathogenesis of psoriasis. In future, it could be useful to use these parameters as non-invasive prognostic biomarkers and new therapeutic targets for psoriasis patients.
Declaration of patient consent
The authors certify that they have obtained all appropriate patient consent.
Financial support and sponsorship
Nil.
Conflict of interest
There are no conflicts of interest.
Glossary of terms
G1P-3: Glucose-Dependent Insulinotropic Polypeptide
3NPM1: Nucleolar Phosphoprotein-1
RNAs: Ribonucleic Acid
IFI6: Interferon Alpha Inducible Protein 6
PRINS: Psoriasis Associated Non-Protein Coding RNA Induced By Stress
UVB: Ultraviolet B
PASI: Psoriasis Area and Severity Index
ND: NanoDrop®
cDNA: complementary Deoxyribonucleic Acid
PCR: Polymerase Chain Reaction
GAPDH: Glyceraldehyde 3-Phosphate Dehydrogenase
SPSS: Statistical Package for the Social Sciences
ROC: Receiver Operating Characteristic
AUC: Area Under the Curve
TGF-β1: Transforming Growth Factor beta
VEGF-A: Vascular Endothelial Growth Factor A
References
- Upregulation of the miRNA-155, miRNA-210, and miRNA-20b in psoriasis patients and their relation to IL-17. Int J Immunopathol Pharmacol. 2020;34 doi:10.1177/2058738420933742
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Serum and tissue expression of transforming growth factor beta 1 in psoriasis. J Eur Acad Dermatol Venereol. 2009;23:406-9.
- [CrossRef] [PubMed] [Google Scholar]
- Expression of sirtuins 1, 6, tumor necrosis factor, and interferon-γ in psoriatic patients. Int J Immunopathol Pharmacol. 2016;29:764-8.
- [CrossRef] [PubMed] [Google Scholar]
- Transcriptome wide analysis of long non-coding RNA-associated ceRNA regulatory circuits in psoriasis. J Cell Mol Med. 2021;25:6925-35.
- [CrossRef] [PubMed] [Google Scholar]
- The anti-apoptotic protein G1P3 is overexpressed in psoriasis and regulated by the non-coding RNA, PRINS. Exp Dermatol. 2010;19:269-78.
- [CrossRef] [PubMed] [Google Scholar]
- PRINS, a primate-specific long non-coding RNA, plays a role in the keratinocyte stress response and psoriasis pathogenesis. Pflugers Arch. 2016;468:935-43.
- [CrossRef] [Google Scholar]
- Expression and functional studies on the noncoding RNA, PRINS. Int J Mol Sci. 2012;14:205-25.
- [CrossRef] [PubMed] [Google Scholar]
- An update on the role of long non-coding RNAs in psoriasis. Chin Med J (Engl). 2020;134:379-89.
- [CrossRef] [PubMed] [Google Scholar]
- Nucleolar phosphoprotein modulates the alleviation of atopic dermatitis caused by the marine-derived compound dihydroaustrasulfone alcohol. Exp Mol Med. 2018;50:e446.
- [CrossRef] [PubMed] [Google Scholar]
- [Features of the structure and expression of NPM and NCL Genes in cutaneous melanoma] Mol Biol (Mosk). 2019;53:663-73.
- [CrossRef] [PubMed] [Google Scholar]
- Clinical measures of disease severity and outcome in psoriasis: A critical appraisal of their quality. Br J Dermatol. 1999;141:185-91.
- [CrossRef] [PubMed] [Google Scholar]
- The eminent roles of ncRNAs in the pathogenesis of psoriasis. Noncoding RNA Res. 2020;5:99-108.
- [CrossRef] [PubMed] [Google Scholar]
- Identification and characterization of a novel, psoriasis susceptibility-related noncoding RNA gene, PRINS. J Biol Chem. 2005;280:24159-67.
- [CrossRef] [PubMed] [Google Scholar]
- Comparison of stress-induced PRINS gene expression in normal human keratinocytes and HaCaT cells. Arch Dermatol Res. 2011;303:745-52.
- [CrossRef] [PubMed] [Google Scholar]
- PRINS lncRNA is a new biomarker candidate for HPV infection and prognosis of head and neck squamous cell carcinomas. Diagnostics (Basel). 2020;10:762.
- [CrossRef] [PubMed] [Google Scholar]
- Long Noncoding RNA Expression in adrenal cortical neoplasms. Endocr Pathol. 2020;31:385-91.
- [CrossRef] [PubMed] [Google Scholar]
- Estrogen stimulates the proliferation of human endometrial cancer cells by stabilizing nucleolar phosphoprotein/B23 (NPM/B23) J Mol Med (Berl). 2013;91:249-59.
- [CrossRef] [PubMed] [Google Scholar]
- Knockdown of nucleolar phosphoprotein induces S-phase arrest in HepG2 cells. Chin J Cancer. 2011;30:853-60.
- [CrossRef] [PubMed] [Google Scholar]
- Antiangiogenic effect of methotrexate and PUVA on psoriasis. Cell Biochem Biophys. 2013;67:735-42.
- [CrossRef] [PubMed] [Google Scholar]






