Translate this page into:
Lack of association of IL-10 (rs1800896) and IL-13 (rs1800925) with non-segmental vitiligo susceptibility in South Indian population
2 Department of Dermatology, Jawaharlal Institute of Postgraduate Medical Education and Research, Puducherry, India
3 Unit of Microbiology and Molecular Biology, Indian Council of Medical Research-Vector Control Research Centre, Puducherry, India
Correspondence Address:
Medha Rajappa
Department of Biochemistry, Jawaharlal Institute of Postgraduate Medical Education and Research, Puducherry - 605 006
India
| How to cite this article: Rajendiran KS, Rajappa M, Chandrashekar L, Thappa D M, Devaraju P. Lack of association of IL-10 (rs1800896) and IL-13 (rs1800925) with non-segmental vitiligo susceptibility in South Indian population. Indian J Dermatol Venereol Leprol 2020;86:489-498 |
Abstract
Background: Vitiligo is an autoimmune depigmentation disorder caused by multiple etiologies. Genetic polymorphisms in cytokine genes influence their expression and augment disease development. Analyzing the influence of genetic polymorphisms will help in better understanding of the complex etiopathogenesis of vitiligo.
Aim: To study the influence of interleukin IL-10 (rs1800896) and IL-13 (rs1800925) polymorphisms on vitiligo risk in South Indian population.
Methods: Two hundred and sixty-four vitiligo patients and 264 controls were recruited in this study. Genotyping was done by quantitative PCR and plasma cytokine levels were measured by ELISA.
Results: Allele frequencies of IL-10 (rs1800896) and IL-13 (rs1800925) SNPs were observed to be equal in the groups. Mutant allele G of IL-10 (rs1800896) enhanced the familial inheritance of vitiligo (P < 0.0001, OR-25.1, 95% CI-7.64–82.7) and influenced the development of vulgaris type of vitiligo (P = 0.034, OR-1.83, 95% CI-1.07–3.13). Ancestral allele A of IL-10 (rs1800896) conferred protection against development of acrofacial vitiligo (P = 0.04, OR-0.56, 95% CI-0.33–0.95). Circulatory IL-10 levels in vitiligo patients were higher than controls (P < 0.0001). Individuals with genotype GG of IL-10 (rs1800896) had the highest circulatory levels of IL-10 (P < 0.0001). Among the genotypes of IL-13 (rs1800925) variant, none influenced the phenotype of nonsegmental vitiligo such as gender, family history, age of onset and types of vitiligo (P > 0.05). In addition, no difference was noted in the circulatory levels of IL-13 between patients and controls (P = 0.48). Within patients, CC genotype of IL-13 (rs1800925) was observed to enhance the circulatory IL-13 levels (P < 0.0001).
Limitation: Replication group analysis in a larger multicentric cohort in future would validate further understanding of vitiligo susceptibility in South Indian ethnics.
Conclusion: IL-10 (rs1800896) and IL-13 (rs1800925) polymorphisms did not confer risk to develop vitiligo in South Indian population.
Introduction
Vitiligo is a skin depigmentation disorder in which the depigmentation occurs due to the damage of functional melanocytes in the epidermal unit. Pathogenesis of vitiligo is very complex and many studies have documented that both genetic and biochemical factors play a key role in determining the disease susceptibility and manifestation of varied clinical phenotypes.[1],[2],[3] In addition, several reports have explained the autoimmune-mediated destruction of melanocytes as the primary cause of the decreased melanin synthesis in vitiligo.[4],[5] Evidence suggests that disturbance in the immune homeostasis and a rise in inflammatory cytokines such as interleukin (IL)-6, IL-8, IL-10, IL-13 and IL-17A contribute to the T cell-mediated inflammatory immune response in vitiligo.[6],[7]
One of the most important immunoregulatory cytokines that has a predominant role in both inflammation and autoimmunity is IL-10.[8] It is secreted by T-helper 2 cells, B cells, mast cells and monocytes/macrophages. The gene IL-10 is located on the chromosome 1.[9] Genetic changes in the upstream and coding regions of cytokine genes are known to alter their circulatory levels, in turn triggering or amplifying autoimmune responses.[10] A previous study on the influence of cytokines in meningococcal disease explicated that nearly 75% of the variation in IL-10 synthesis is determined by genetic variants.[11] Several studies on autoimmune diseases such as rheumatoid arthritis (RA), systemic lupus erythematosus (SLE) and autoimmune thyroid disease have reported the up-regulation of IL-10 along with transforming growth factor, TGF-β.[8],[12],[13],[14] Experiments conducted on an animal model for human vitiligo suggest that there is a Th-1polarized autoimmunity in vitiligo, with increased expression of IFN-γ and IL-10 in active disease, suggesting the influence of genetic changes in these cytokine variants with vitiligo development.[15] Single nucleotide polymorphisms (SNP) in the promoter region of the IL-10 gene influence the expression of IL-10.[16],[17],[18],[19] Among the several SNPs of IL-10, the variant at -1082(A/G), rs1800896 is recognized to be strongly associated with autoimmune diseases such as RA, SLE and autoimmune thyroid disease.[20],[21],[22],[23] A few studies on vitiligo susceptibility from other ethnicities have reported that the G allele of IL-10 (rs1800896) is associated with vitiligo risk and increased serum IL-10 level in vitiligo.[24],[25]
Likewise, another antiinflammatory cytokine known to share similarity with IL-4 is IL-13, which is excessively secreted by activated T helper cells and dendritic cells. It mediates the regulation of pro-inflammatory cytokines and MHC class II (MHC II) expression by monocytes.[26],[27],[28] This antiinflammatory cytokine plays a pivotal role in the pathogenesis of several diseases such as asthma, autoimmune thyroid disease, RA, SLE, eczema, pulmonary disease and atopy through hypersensitivity, IgE synthesis, mast cell hyperplasia, tissue remodeling and fibrosis.[29],[30],[31],[32],[33],[34] The gene encoding for IL-13 is located on chromosome 5q31–33. Recent research has revealed that this location is one of the vulnerable regions for the susceptibility of asthma or atopic dermatitis, which suggests that IL-13 could have an association with autoimmune diseases.[35] Experimental evidence suggests that the most common SNP of the IL-13 gene, rs1800925 (–1055C/T), also called –1112C/T is associated with increased levels of IL-13 from activated T cells in allergy.[29],[36] However, the genetic association of IL-13 variants with vitiligo risk has not been established till date. Alteration in the immune homeostasis in vitiligo and an elevation of serum IL-13 reported in vitiligo patients suggest that it plays a role in the pathogenesis of disease progression.[6]
General occurrence of skin-related disorders may vary due to differences in allele variations of the genes among diverse populations and ethnicities.[37],[38] Given the background of Th2 cytokine gene variants in the autoimmune diseases and the genetic association of skin diseases, we initiated this study as a preliminary work to check for association of the two most common SNPs from Th2 cytokines, IL-10 (rs1800896, A/G) and IL-13 (rs1800925, C/T), on vitiligo susceptibility in South Indian Tamil population. This would help to extend our knowledge on the genetic risk factors associated with vitiligo susceptibility and their influence on the disease phenotypes.
Methods
Study subjects
Two hundred and sixty-four patients of Tamil ethnicity suffering from vitiligo[39] who presented to the Dermatology Clinic at Jawaharlal Institute of Postgraduate Medical Education and Research (JIPMER), a tertiary care hospital located in Puducherry were recruited as cases in this research, as a part of a larger hospital-based cohort study.[40] A questionnaire was used to record demographic details such as disease type, onset age, duration and family history of autoimmune disease. Disease activity was assessed by vitiligo index of disease activity (VIDA) scoring.[41] Two hundred and sixty-four age, gender and ethnicity-matched individuals without dermatological disorders, malignancies, infectious diseases and family history of autoimmune diseases were enrolled as controls. The study was conducted as per the World Medical Association Declaration of Helsinki ethical principles for medical research involving human subjects. JIPMER Institute Ethics Committee for Human studies approved the study. The study procedure was well explained and written informed consent was obtained from all the participants.
Genotyping
Five milliliters of the blood sample was collected from the study subjects. Plasma was separated and stored at −40°C for quantification of cytokines. Modified salting-out method was used to isolate genomic DNA from the blood.[42] DNA was quantified (Nano Drop Technologies Inc., USA) and diluted to contain 100 ng/μl and used for genotyping. IL-10 (rs1800896) and IL-13 (rs1800925) genetic variants were genotyped using TaqMan Allelic Discrimination Assay by using reagents from Applied Biosystems, California, USA with assay IDs: C__1747362_10 and C__8932056_10 on a Bio-Rad CFX Real-time detection system (Bio-Rad CFX, California, USA), following the protocols of the manufacturer. Replicates of thirty per cent of random samples were genotyped to validate genotyping results and to rule out the technical or observational error.
Cytokine assay
Circulating plasma levels IL-10 and IL-13 in all the study subjects were measured using commercially available ELISA kits (Diaclone Research, France, Cat. No: 950.060.096 - human IL-10 ELISA kit Cat. No: 850.080.096 - human IL-13 ELISA kit).
Sample size calculation
The sample size was computed using CaTS Power Calculator Software (Power Calculator for Genetic Studies, Centre for Statistical Genetics, Michigan University, USA). A sample size of 220 cases and 220 controls was estimated at 5% level of significance with 80% power for the estimated disease prevalence of 2.5% and minor allele frequency of 24.7% for IL-13 (rs1800925) (https://www.ncbi.nlm.nih.gov/projects/SNP/snp_ss.cgi?ss=ss65831236).[43] The sample size was further revised to 264 subjects in each group for empowering an analysis of different traits.
Statistical analysis
Direct gene counting was carried out to identify the genotype frequencies in both cases and controls. The observed genotype frequencies in cases and controls were tested for Hardy–Weinberg equilibrium (HWE) by Chi-square test. The genotype and allele frequencies in cases and controls were compared using the Chi-square test. Normality of the data was assessed by the Kolmogorov–Smirnov test. Mann–Whitney U Test was used to compare cytokine levels between cases and controls. Kruskal–Wallis test was used to compare cytokine levels of the respective genotypes of SNPs. The analysis was done at 5% level of significance and a two-sided P < 0.05 was considered as significant. However, for multiple pairwise comparisons, P < 0.005 was considered statistically significant. All the statistical analysis and graphs were made using GraphPad Prism 5 (Graph Pad Software, Inc., United States of America).
Results
Demographic and clinical characteristics of the study subjects
The baseline characteristics of the study population have been published previously[40] and are given in [Table - 1]. Mean age of patients at the time of enrolment was 40.34 ± 14.27 years and the observed mean age of onset was 33.58 ± 15.80 years.[40] The observed male:female ratio was equal (1:1) in both the groups. At the time of recruitment, the median VIDA in vitiligo patients was 2 (1-3).

Genotyping results
The observed ancestral allele frequency of IL-10 (rs1800896) polymorphism in cases and control group was 342 (64.8%) and 340 (64.4%), respectively. No significant difference in the frequencies of mutant allele G (rs1800896) was observed between cases and controls (35.2% in cases and 35.6% in controls). Statistical analysis revealed that the mutant allele G was not a genetic risk factor to develop vitiligo in south Indian population (P = 0.95) [Table - 2]. It was also observed that inheritance of the heterozygous AG genotype of IL-10 (rs1800896) polymorphism among family members enhances the risk to develop vitiligo (P < 0.0001, OR-25.1, 95% CI-7.64–82.7). However, in cases, the heterozygous AG genotype of IL-10 (rs1800896) polymorphism was significantly associated with the development of vulgaris type (P = 0.034, OR-1.83, 95% CI-1.07–3.13). In addition, by dominant model of genetic analysis, it was observed that the ancestral allele A of IL-10 (rs1800896) polymorphism confers protection against the development of acrofacial type (P = 0.04, OR-0.56, 95% CI-0.33–0.95). However, after adjusting for multiple testing, we did not observe any statistically significant difference for types of vitiligo with this SNP (P > 0.005). We did not find any significant differences in the other subphenotypes such as the age of onset, gender and disease activity [Table - 3]. For the genetic variant, IL-13 (rs1800925), no significant association with the development of the disease was observed (P = 0.95) [Table - 4]. We observed that the IL-13 (rs1800925) polymorphism neither conferred significant risk to develop the disease nor influenced the subphenotypes in south Indian population (P > 0.005) [Table - 5].
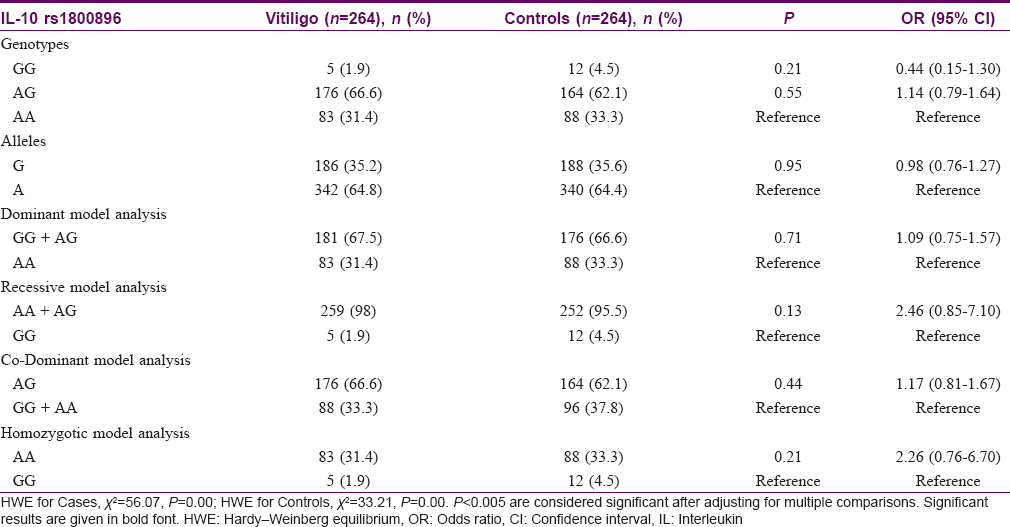
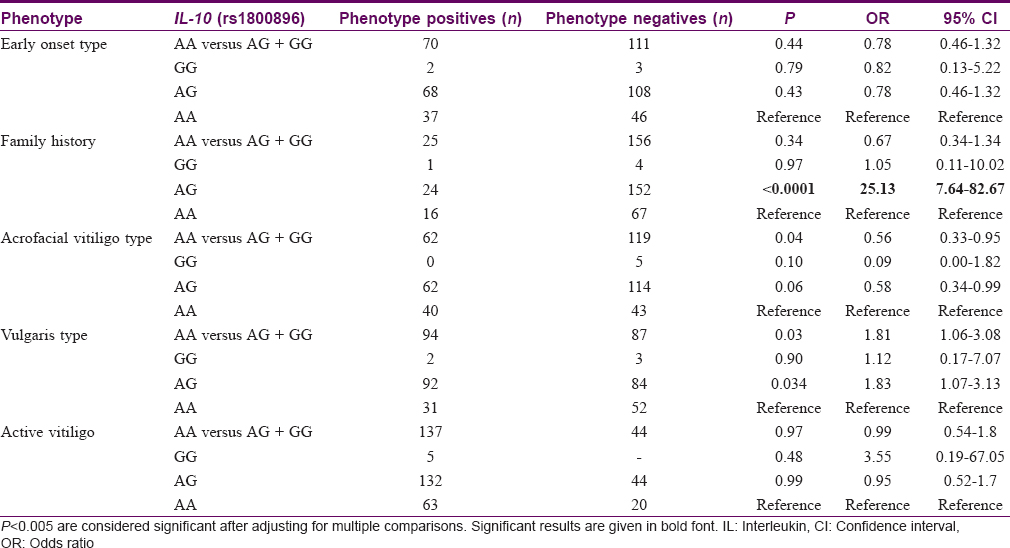

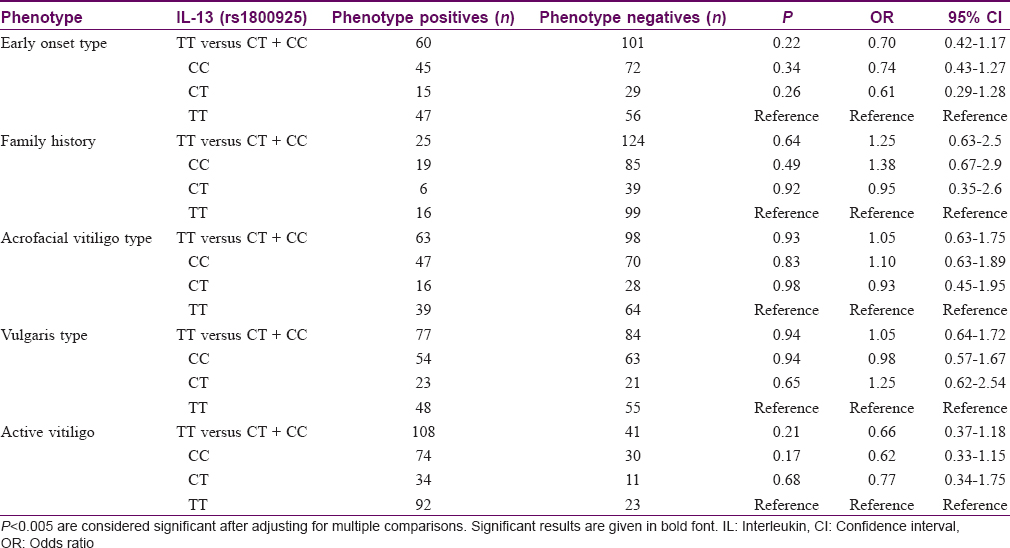
Influence of genotypes on circulating the levels of the cytokines
Comparison of median IL-10 plasma levels between cases and controls revealed that the circulatory concentration of IL-10 was significantly higher in cases than in controls (P < 0.0001) [Figure - 1]. However, no significant difference in the circulatory levels of IL-13 was observed between the cases and controls (P = 0.48) [Figure - 2]. Analysis of th influence of polymorphism on circulatory levels of cytokines indicated that the patients harboring IL-10 (rs1800896) homozygous variant GG genotype [Median, (IQR): 45.02 (35.95–82.44) pg/mL] had higher IL-10 plasma concentration than the patients carrying AG [Median, (IQR): 12.46 (8.5–17.74) pg/mL] with P = 0.001, and AA genotype [Median, (IQR): 7.76 (5.21–13.35) pg/mL] genotypes with P = 0.001 [Figure - 3]. Similarly, the individuals with homozygous variant CC of IL-13(rs1800925) [Median, (IQR): 22.81 (11.91–45.73) had comparable levels of IL-13 in individuals with heterozygous CT [Median, (IQR): 20.57 (10.13–46.47) pg/mL, (P = 0.45)], whereas increased levels of IL-13 compared to the individuals with TT genotypes [Median, (IQR):11.30 (5.59–17.66) pg/mL, (P < 0.0001)] [Figure - 4].
 |
| Figure 1: Illustrates the plasma IL-10 levels in patients with cases (n = 264) and controls (n = 264) median, (IQR): 13.27 (8.27–23.28) versus 6.80 (5.26–11.96) pg/mL, P < 0.0001. IQR: Interquartile range, IL: Interleukin |
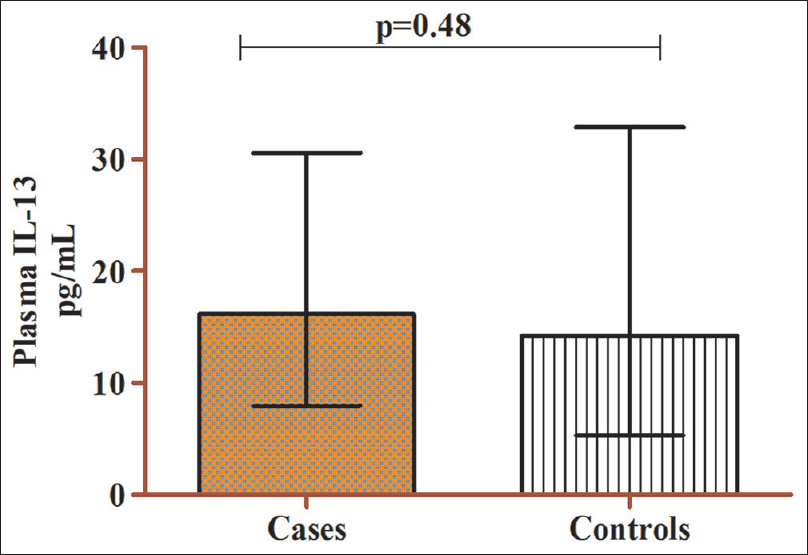 |
| Figure 2: Depicts plasma IL-13 levels in patients with cases (n = 264) and controls (n = 264) median, (IQR): 16.15 (7.93–30.59) versus 14.25 (5.30–32.90) pg/mL, P = 0.48. IQR: Interquartile range, IL: Interleukin |
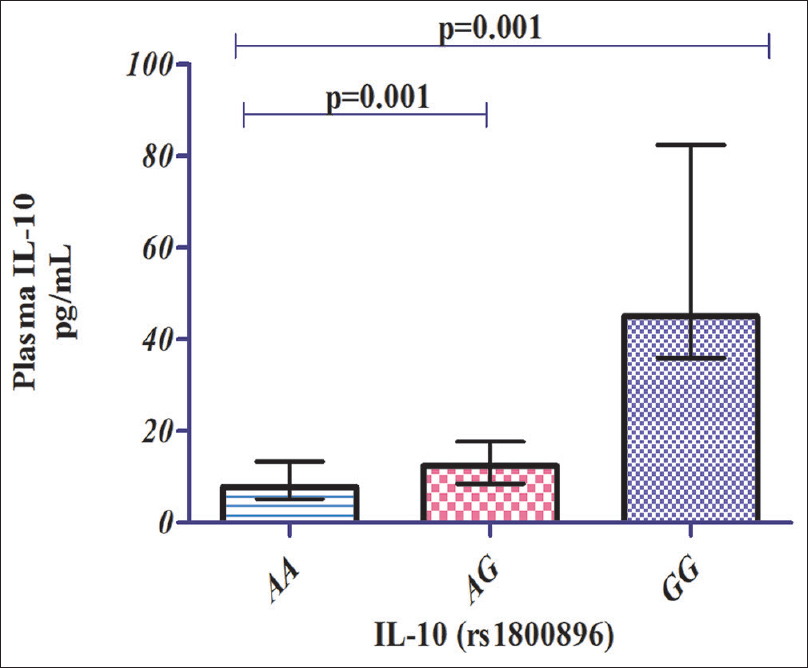 |
| Figure 3: Depicts the influence of IL-10 (rs1800896) genotypes on plasma IL-10 levels in cases (n = 264) Wild AA (n = 83) median, (IQR): 7.76 (5.21–13.35) versus heterozygous AG (n = 176) median, (IQR): 12.46 (8.5–17.74) pg/mL, P = 0.001. Wild AA (n = 83) median, (IQR): 7.76 (5.21–13.35) versus homozygous mutant GG (n = 5) median, (IQR): 45.02 (35.95–82.44) pg/mL, P = 0.001. IQR: Interquartile range, IL: Interleukin |
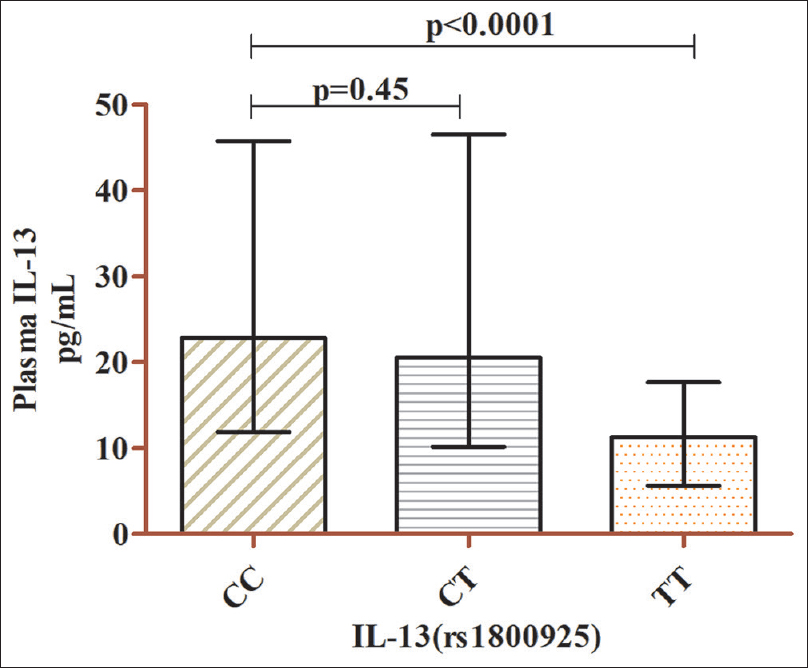 |
| Figure 4: Depicts the influence of IL-13 (rs1800925) genotypes on plasma IL-13 levels in cases (n = 264) Wild CC (n = 115) median, (IQR): 22.81 (11.91–45.73) versus heterozygous CT (n = 45) median, (IQR): 20.57 (10.13–46.47) pg/mL, P = 0.45. Wild CC (n = 115) median, (IQR): 22.81 (11.91–45.73) versus homozygous mutant TT (n = 104) median, (IQR): 11.30 (5.59–17.66) pg/mL, P < 0.0001. IQR: Interquartile range, IL: Interleukin |
Correlation of plasma Th2 cytokine levels with vitiligo index of disease activity scores
Spearman correlation analysis was performed to study the correlation of circulatory IL-10/IL-13 cytokine levels with VIDA score (r = 0.018; P = 0.76 and r = 0.016; P = 0.79, respectively), which revealed that there was no statistically significant correlation between the variables [Figure - 5].
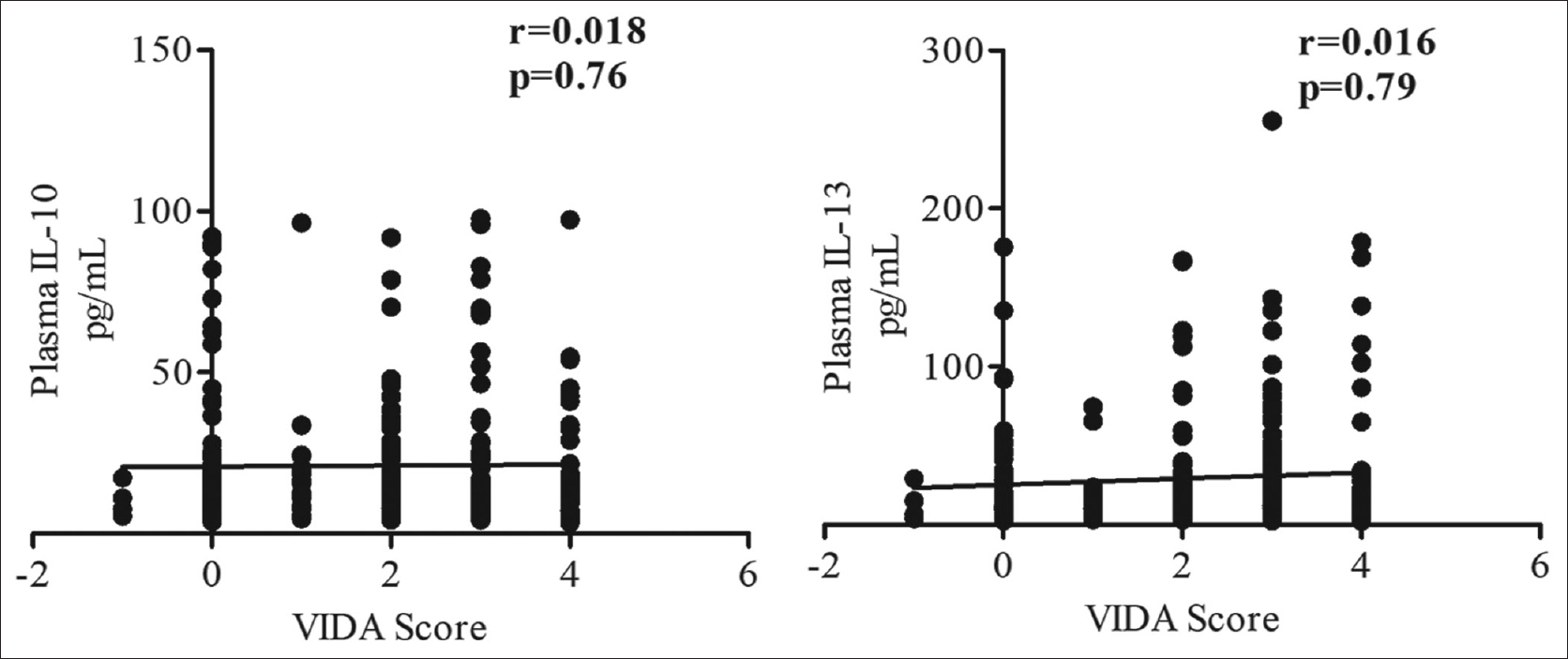 |
| Figure 5: Depicts the correlation between VIDA score and plasma IL-10 and IL-13 levels in all cases (n = 264). VIDA: Vitiligo index of disease activity |
Discussion
Vitiligo is an autoimmune skin disease with genetic predisposition as a key etiologic factor. In autoimmune disease, cytokine surge favors a break in immune tolerance and amplification of autoimmune responses.[44] When a break in immune tolerance occurs, the cytokine secretions by T cell subsets are invariably affected. In this complex autoimmune process, IL-10, though anti-inflammatory by nature, is known to play a pathogenic role in the development of the vitiligo.[44],[45] IL-13, a Th2 anti-inflammatory cytokine, which was once believed to have a significant role in allergy, is now known to mediate anti-inflammatory processes in susceptibility and progression of various autoimmune diseases.[46] Previous studies have demonstrated increased expression of inflammatory cytokines in skin and circulation among vitiligo patients.[47],[48]
In this study, we explored the association of IL-10 (rs1800896) and IL-13 (rs1800925) polymorphisms with the development of vitiligo and its associated clinical phenotype. The frequency of ancestral A allele of IL-10 (rs1800896) variant in our control group was 340 (64.4%), which is slightly lesser than the frequency 422 (69%) reported in a North Indian cohort[49] and much higher than the frequency reported in Saudi population 110 (54.5%).[24] No such deviation was observed in comparison with a Turkish 269 (64%) population.[25] However, analysis of vitiligo in our study population revealed that the minor variant of IL-10 (rs1800896) did not confer risk to develop vitiligo. Our findings are in contrast with the reports from studies conducted in a Turkish and Saudi cohort, in whom the minor variant was reported to be a genetic risk factor to develop vitiligo. The observed difference in the susceptibility between the different populations can be attributed to the genetic heterogeneity of different ethnic groups and environmental factors.[38]
Although the IL-10 (rs1800896) polymorphism was not found to be a genetic risk factor in our population, we found that the inheritance of a single copy of the mutant allele G enhanced vitiligo risk in siblings. We observed a lack of familial inheritance in individuals harboring GG genotype, which could be due to the lesser occurrence of the double mutant in our study groups. We also observed that the individuals with AG genotype showed higher propensity to develop vulgaris type of vitiligo and the ancestral allele A conferred protection against the development of acrofacial vitiligo. However, on adjustment for multiple comparison, IL-10 (rs1800896) was not significantly associated with type of vitiligo (P > 0.005). The observed frequency of IL-10 genotypes in our population was not concordant with HWE. Deviation from HWE in this study could be due to the practice of endogamous marriage in this population and increased admixture of population.[50],[51],[52],[53] Deviation due to genotyping observational error was ruled out by repeating the analysis in 30% randomly selected samples.
Influence of the genetic polymorphism of IL-10 (rs1800896) on the expression of the cytokine revealed that the mutant allele G significantly enhanced the secretion of IL-10. Our result is in line with other earlier reports demonstrating increased levels in vitiligo in individuals with GG genotype.[24],[25] A study in multiple sclerosis showed similar results where increased IL-10 producer GG genotype and decreased producer AA genotypes had no association with the disease.[54] Correspondingly, Stanilova showed that GG genotype is associated with increased IL-10 production in severe sepsis, which is in concordance with our results.[55] Also in line with this, a case-control study on pneumonia showed that Egyptian children with GG genotype had increased levels of IL-10.[56] This increased circulatory IL-10 level of individuals with a single copy or double copy of the mutant allele compared to individuals with homozygous ancestral genotype AA in our study suggests that a single copy of the mutant allele G enhances the IL-10 expression. Our findings are in tandem with the earlier reports on influence of -1082G IL-10 (rs1800896) polymorphism on circulatory IL-10 levels.[57]
Increased IL-10 levels mediate humoral immune mechanisms in inflammatory conditions by increasing the production of immunoglobulin such as IgG, IgA and IgM from activated B cells.[58] Thus, by antibody production, IL-10 augments melanocyte damage in vitiligo pathogenesis. A rise in IL-10 levels in conjunction with a prevailing autoimmune response probably augments the pro-inflammatory responses via IFN-γ, thereby moderating the vitiligo pathogenesis and/or its clinical phenotypes.[15] Ala et al. showed that IL-10 was decreased in vitiligo patients, while IFN-γ:IL-10 ratio was an important marker in nonsegmental vitiligo. However, their study was limited by a small number of samples.[59] Grimes et al. reported that after tacrolimus therapy, vitiligo patients showed no significant change in IL-10 levels.[60]
During inflammation, T cell infiltration is counter-regulated by IL-10, an anti-inflammatory cytokine secreted by regulatory T cells, which are documented to regulate T cell proliferation and cytokine synthesis. The probable increase could be due to the effect of anti-inflammatory response, which could not counter regulate the pro-inflammatory action in the lesion causing vitiligo progression. In line with this theory, Taher et al. showed increased elevation of IL-10 levels with mild follicular repigmentation in vitiligo patients after treatment with tacrolimus.[61]
In view of the IL-13 gene emerging as a promising therapeutic target for autoimmune diseases, we tested the influence of IL-13 on vitiligo development and its clinical manifestations and found that the IL-13 promoter region polymorphism (rs1800925) was not a genetic risk factor for vitiligo in our population.[62],[63],[64] In our study, equal distribution of wild and ancestral alleles in cases and control groups rendered this polymorphism to be a poor genetic risk factor for vitiligo. In support of our results, Chong et al. reported that IL-13 was not associated with risk of Grave's disease in Chinese population with P = 0.22 after Bonferroni's correction. However, they insisted on the use of large sample size and different ethnicity for further confirmation.[64] A recent meta-analysis on allergic rhinitis demonstrated no significant association of –1112C/T (rs1800925) with disease development.[65]
In contrast, in autoimmune diseases such as psoriatic arthritis, the authors observed that IL-13 gene polymorphism was strongly associated with increased odds ratio of 1.28 and observed that smoking was found to alter the association of –1112C/T (rs1800925) with disease susceptibility.[63],[66] A recent meta-analysis demonstrated a strong T allele association of –1112C/T polymorphism with increased risk of COPD in Asian and Caucasian population.[67] Pertovaara et al. revealed strong association of IL-13 (rs1800925) with the mild type of primary Sjogren's syndrome.[68] In our research, comparison of median circulatory levels of IL-13 in controls and cases revealed a slight but statistically insignificant rise of IL-13 in cases. The IL-13 exerts its anti-inflammatory activity by stimulating the macrophages to secrete TGF-β.[69] von Willebrand et al. had reported that the TGF-β induces apoptosis in cultured normal melanocytes and inhibits melanocyte activity.[70] Though we did not find IL-13 (rs1800925) to be associated with vitiligo risk in our population, we hypothesize that IL-13 could contribute to vitiligo pathogenesis by favoring melanocyte destruction or down-regulation of melanocyte activity through TGF-β. In addition, TGF-β might break immune tolerance and would favor autoimmune-mediated destruction of melanocytes in vitiligo. This warrants a future study on combined effect of IL-13 and TGF-β in vitiligo pathogenesis.
Comparable levels of IL-13 in the major homozygous CC and heterozygous CT genotypes of cases suggested that dominant allele C is expressed, excluding the influence of T allele in CT (P = 0.45). However, no positive correlation between IL-13 (rs1800925) with disease susceptibility and association with clinical phenotype was observed in our study. Cameron et al. have stated that the T allele at position –1112 C/T may amplify binding of the NFAT protein to this promoter region of IL-13 gene.[36] van der Pouw Kraan et al. reported that the mutant T allele was associated with upregulation of IL-13 expression. Their experiments demonstrated that the change of allele C to T increased the binding of nuclear factor of activated T cells (NFAT), thereby accounting for the elevated expression of IL-13.[29] In systemic sclerosis, Broen et al. demonstrated that IL-13 (rs1800925) was not associated with disease susceptibiltiy and does not have an effect on their mRNA expression in blood.[71] In our study, CC genotype showed elevated IL-13 levels than mutant TT genotype, which could be due to suppression of T allele in vitiligo. Increased cytokines levels in vitiligo may be due to immune activation mechanisms that promote Th-2 differentiation of innate immune cells.[72]
Study of several other SNPs of these cytokine genes would have increased the understanding of the influence of the linkage disequilibrium with the vitiligo susceptibility, which is a limitation of our study.
Conclusion
The equal distribution of IL-10 (rs1800896) and IL-13 (rs1800925) frequencies in our study population rendered these variants a non-risk factor for vitiligo in South Indian population. However, the IL-10 (rs1800896) polymorphism was observed to influence the circulatory levels of IL-10 in vitiligo patients and enhanced the familial inheritance of vitiligo. Replication group analysis in a larger multicentric cohort in the future would validate further understanding of vitiligo susceptibility in South Indian ethnics.
Acknowledgement
We are thankful to the patients and the control volunteers for their participation in the research. We are grateful to JIPMER for intramural grant in the name of the corresponding author for this research.
Declaration of patient consent
The authors certify that they have obtained all appropriate patient consent forms. In the form, the patients have given their consent for their images and other clinical information to be reported in the journal. The patients understand that their names and initials will not be published and due efforts will be made to conceal their identity, but anonymity cannot be guaranteed.
Financial support and sponsorship
Intramural research grant from Jawaharlal Institute of Post Graduate Medical Education and Research, Puducherry, India, in the name of the corresponding author is gratefully acknowledged.
Conflicts of interest
There are no conflicts of interest.
| 1. |
Al-Shobaili HA. Update on the genetics characterization of Vitiligo. Int J Health Sci 2011;5:167-79.
[Google Scholar]
|
| 2. |
Spritz RA. The genetics of vitiligo. J Invest Dermatol 2011;131:E18-20.
[Google Scholar]
|
| 3. |
Furue M, Kadono T. Nonsegmental vitiligo update. Dermatol Sin 2016;34:174-6.
[Google Scholar]
|
| 4. |
Sandoval-Cruz M, García-Carrasco M, Sánchez-Porras R, Mendoza-Pinto C, Jiménez-Hernández M, Munguía-Realpozo P, et al. Immunopathogenesis of vitiligo. Autoimmun Rev 2011;10:762-5.
[Google Scholar]
|
| 5. |
Boniface K, Seneschal J, Picardo M, Taïeb A. Vitiligo: Focus on clinical aspects, immunopathogenesis, and therapy. Clin Rev Allergy Immunol 2018;54:52-67.
[Google Scholar]
|
| 6. |
Tembhre MK, Sharma VK, Sharma A, Chattopadhyay P, Gupta S. T helper and regulatory T cell cytokine profile in active, stable and narrow band ultraviolet B treated generalized vitiligo. Clin Chim Acta 2013;424:27-32.
[Google Scholar]
|
| 7. |
Moretti S, Spallanzani A, Amato L, Hautmann G, Gallerani I, Fabiani M, et al. New insights into the pathogenesis of vitiligo: Imbalance of epidermal cytokines at sites of lesions. Pigment Cell Res 2002;15:87-92.
[Google Scholar]
|
| 8. |
Bidwell J, Keen L, Gallagher G, Kimberly R, Huizinga T, McDermott MF, et al. Cytokine gene polymorphism in human disease: On-line databases. Genes Immun 1999;1:3-19.
[Google Scholar]
|
| 9. |
Lalani I, Bhol K, Ahmed AR. Interleukin-10: Biology, role in inflammation and autoimmunity. Ann Allergy Asthma Immunol 1997;79:469-83.
[Google Scholar]
|
| 10. |
Kim JM, Brannan CI, Copeland NG, Jenkins NA, Khan TA, Moore KW. Structure of the mouse IL-10 gene and chromosomal localization of the mouse and human genes. J Immunol 1992;148:3618-23.
[Google Scholar]
|
| 11. |
Westendorp RG, Langermans JA, Huizinga TW, Elouali AH, Verweij CL, Boomsma DI, et al. Genetic influence on cytokine production and fatal meningococcal disease. Lancet 1997;349:170-3.
[Google Scholar]
|
| 12. |
Bucht A, Larsson P, Weisbrot L, Thorne C, Pisa P, Smedegård G, et al. Expression of interferon-gamma (IFN-gamma), IL-10, IL-12 and transforming growth factor-beta (TGF-beta) mRNA in synovial fluid cells from patients in the early and late phases of rheumatoid arthritis (RA). Clin Exp Immunol 1996;103:357-67.
[Google Scholar]
|
| 13. |
Feldmann M, Brennan FM, Maini RN. Role of cytokines in rheumatoid arthritis. Annu Rev Immunol 1996;14:397-440.
[Google Scholar]
|
| 14. |
Katsikis PD, Chu CQ, Brennan FM, Maini RN, Feldmann M. Immunoregulatory role of interleukin 10 in rheumatoid arthritis. J Exp Med 1994;179:1517-27.
[Google Scholar]
|
| 15. |
Shi F, Erf GF. IFN-γ, IL-21, and IL-10 co-expression in evolving autoimmune vitiligo lesions of Smyth line chickens. J Invest Dermatol 2012;132:642-9.
[Google Scholar]
|
| 16. |
Eskdale J, Gallagher G, Verweij CL, Keijsers V, Westendorp RG, Huizinga TW. Interleukin 10 secretion in relation to human IL-10 locus haplotypes. Proc Natl Acad Sci U S A 1998;95:9465-70.
[Google Scholar]
|
| 17. |
Mörmann M, Rieth H, Hua TD, Assohou C, Roupelieva M, Hu SL, et al. Mosaics of gene variations in the interleukin-10 gene promoter affect interleukin-10 production depending on the stimulation used. Genes Immun 2004;5:246-55.
[Google Scholar]
|
| 18. |
Turner DM, Williams DM, Sankaran D, Lazarus M, Sinnott PJ, Hutchinson IV. An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet 1997;24:1-8.
[Google Scholar]
|
| 19. |
Burdin N, Péronne C, Banchereau J, Rousset F. Epstein-Barr virus transformation induces B lymphocytes to produce human interleukin 10. J Exp Med 1993;177:295-304.
[Google Scholar]
|
| 20. |
Godsell J, Rudloff I, Kandane-Rathnayake R, Hoi A, Nold MF, Morand EF, et al. Clinical associations of IL-10 and IL-37 in systemic lupus erythematosus. Sci Rep 2016;6:34604.
[Google Scholar]
|
| 21. |
Gambhir D, Lawrence A, Aggarwal A, Misra R, Mandal SK, Naik S. Association of tumor necrosis factor alpha and IL-10 promoter polymorphisms with rheumatoid arthritis in North Indian population. Rheumatol Int 2010;30:1211-7.
[Google Scholar]
|
| 22. |
Jung JH, Song GG, Kim JH, Choi SJ. Association of interleukin 10 gene polymorphisms with autoimmune thyroid disease: Meta-analysis. Scand J Immunol 2016;84:272-7.
[Google Scholar]
|
| 23. |
Iyer SS, Cheng G. Role of interleukin 10 transcriptional regulation in inflammation and autoimmune disease. Crit Rev Immunol 2012;32:23-63.
[Google Scholar]
|
| 24. |
Abanmi A, Al Harthi F, Zouman A, Kudwah A, Jamal MA, Arfin M, et al. Association of interleukin-10 gene promoter polymorphisms in Saudi patients with vitiligo. Dis Markers 2008;24:51-7.
[Google Scholar]
|
| 25. |
Aydıngöz IE, Kanmaz-Özer M, Gedikbaşi A, Vural P, Doǧru-Abbasoǧlu S, Uysal M. The combination of tumour necrosis factor-α -308A and interleukin-10 -1082G gene polymorphisms and increased serum levels of related cytokines: Susceptibility to vitiligo. Clin Exp Dermatol 2015;40:71-7.
[Google Scholar]
|
| 26. |
de Waal Malefyt R, Figdor CG, Huijbens R, Mohan-Peterson S, Bennett B, Culpepper J, et al. Effects of IL-13 on phenotype, cytokine production, and cytotoxic function of human monocytes. Comparison with IL-4 and modulation by IFN-gamma or IL-10. J Immunol 1993;151:6370-81.
[Google Scholar]
|
| 27. |
de Vries JE. The role of IL-13 and its receptor in allergy and inflammatory responses. J Allergy Clin Immunol 1998;102:165-9.
[Google Scholar]
|
| 28. |
McKenzie GJ, Bancroft A, Grencis RK, McKenzie AN. A distinct role for interleukin-13 in th2-cell-mediated immune responses. Curr Biol 1998;8:339-42.
[Google Scholar]
|
| 29. |
van der Pouw Kraan TC, van Veen A, Boeije LC, van Tuyl SA, de Groot ER, Stapel SO, et al. An IL-13 promoter polymorphism associated with increased risk of allergic asthma. Genes Immun 1999;1:61-5.
[Google Scholar]
|
| 30. |
Inoue N, Watanabe M, Morita M, Tatusmi K, Hidaka Y, Akamizu T, et al. Association of functional polymorphisms in promoter regions of IL5, IL6 and IL13 genes with development and prognosis of autoimmune thyroid diseases. Clin Exp Immunol 2011;163:318-23.
[Google Scholar]
|
| 31. |
Miyake Y, Tanaka K, Arakawa M. IL13 genetic polymorphisms, smoking, and eczema in women: A case-control study in Japan. BMC Med Genet 2011;12:142.
[Google Scholar]
|
| 32. |
Heinzmann A, Jerkic SP, Ganter K, Kurz T, Blattmann S, Schuchmann L, et al. Association study of the IL13 variant arg110Gln in atopic diseases and juvenile idiopathic arthritis. J Allergy Clin Immunol 2003;112:735-9.
[Google Scholar]
|
| 33. |
Rinaldi T, Spadaro A, Riccieri V, Taccari E, Valesini G. Interleukin-13 (IL-13) in autoimmune rheumatic diseases: Relationship with autoantibody profile. Arthritis Res 2001;3 Suppl A: P022.
[Google Scholar]
|
| 34. |
van der Pouw Kraan TC, Küçükaycan M, Bakker AM, Baggen JM, van der Zee JS, Dentener MA, et al. Chronic obstructive pulmonary disease is associated with the-1055 IL-13 promoter polymorphism. Genes Immun 2002;3:436-9.
[Google Scholar]
|
| 35. |
Cookson WO, Moffatt MF. Genetics of asthma and allergic disease. Hum Mol Genet 2000;9:2359-64.
[Google Scholar]
|
| 36. |
Cameron L, Webster RB, Strempel JM, Kiesler P, Kabesch M, Ramachandran H, et al. Th2 cell-selective enhancement of human IL13 transcription by IL13-1112C and gt; T, a polymorphism associated with allergic inflammation. J Immunol 2006;177:8633-42.
[Google Scholar]
|
| 37. |
Karter AJ. Commentary: Race, genetics, and disease – in search of a middle ground. Int J Epidemiol 2003;32:26-8.
[Google Scholar]
|
| 38. |
Huang T, Shu Y, Cai YD. Genetic differences among ethnic groups. BMC Genomics 2015;16:1093.
[Google Scholar]
|
| 39. |
Gawkrodger DJ, Ormerod AD, Shaw L, Mauri-Sole I, Whitton ME, Watts MJ, et al. Guideline for the diagnosis and management of vitiligo. Br J Dermatol 2008;159:1051-76.
[Google Scholar]
|
| 40. |
Rajendiran KS, Rajappa M, Chandrashekar L, Thappa DM. Association of PTPN22 gene polymorphism with non-segmental vitiligo in South Indian Tamils. Postepy Dermatol Alergol 2018;35:280-5.
[Google Scholar]
|
| 41. |
Njoo MD, Das PK, Bos JD, Westerhof W. Association of the köbner phenomenon with disease activity and therapeutic responsiveness in vitiligo vulgaris. Arch Dermatol 1999;135:407-13.
[Google Scholar]
|
| 42. |
Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988;16:1215.
[Google Scholar]
|
| 43. |
Shajil EM, Chatterjee S, Agrawal D, Bagchi T, Begum R. Vitiligo: Pathomechanisms and genetic polymorphism of susceptible genes. Indian J Exp Biol 2006;44:526-39.
[Google Scholar]
|
| 44. |
Moudgil KD, Choubey D. Cytokines in autoimmunity: Role in induction, regulation, and treatment. J Interferon Cytokine Res 2011;31:695-703.
[Google Scholar]
|
| 45. |
Singh M, Kotnis A, Jadeja SD, Mondal A, Mansuri MS, Begum R. Cytokines: The yin and yang of vitiligo pathogenesis. Expert Rev Clin Immunol 2019;15:177-88.
[Google Scholar]
|
| 46. |
Mao YM, Zhao CN, Leng J, Leng RX, Ye DQ, Zheng SG, et al. Interleukin-13: A promising therapeutic target for autoimmune disease. Cytokine Growth Factor Rev 2019;45:9-23.
[Google Scholar]
|
| 47. |
Sharquie KE, Mehenna SH, Naji AA, Al-Azzawi H. Inflammatory changes in vitiligo: Stage I and II depigmentation. Am J Dermatopathol 2004;26:108-12.
[Google Scholar]
|
| 48. |
Birol A, Kisa U, Kurtipek GS, Kara F, Kocak M, Erkek E, et al. Increased tumor necrosis factor alpha (TNF-alpha) and interleukin 1 alpha (IL1-alpha) levels in the lesional skin of patients with nonsegmental vitiligo. Int J Dermatol 2006;45:992-3.
[Google Scholar]
|
| 49. |
Chatterjee R, Batra J, Kumar A, Mabalirajan U, Nahid S, Niphadkar PV, et al. Interleukin-10 promoter polymorphisms and atopic asthma in North Indians. Clin Exp Allergy 2005;35:914-9.
[Google Scholar]
|
| 50. |
Cockerham CC. Variance of gene frequencies. Evolution 1969;23:72-84.
[Google Scholar]
|
| 51. |
Cockerham CC. Group inbreeding and coancestry. Genetics 1967;56:89-104.
[Google Scholar]
|
| 52. |
Weir BS, Hill WG, Cardon LR, SNP Consortium. Allelic association patterns for a dense SNP map. Genet Epidemiol 2004;27:442-50.
[Google Scholar]
|
| 53. |
Robertson A, Hill WG. Deviations from hardy-weinberg proportions: Sampling variances and use in estimation of inbreeding coefficients. Genetics 1984;107:703-18.
[Google Scholar]
|
| 54. |
Luomala M, Lehtimäki T, Huhtala H, Ukkonen M, Koivula T, Hurme M, et al. Promoter polymorphism of IL-10 and severity of multiple sclerosis. Acta Neurol Scand 2003;108:396-400.
[Google Scholar]
|
| 55. |
Stanilova SA. Functional relevance of IL-10 promoter polymorphisms for sepsis development. Crit Care 2010;14:119.
[Google Scholar]
|
| 56. |
Azab SF, Abdalhady MA, Elsaadany HF, Elkomi MA, Elhindawy EM, Sarhan DT, et al. Interleukin-10 -1082 G/A gene polymorphisms in Egyptian children with CAP: A case-control study. Medicine (Baltimore) 2016;95:e4013.
[Google Scholar]
|
| 57. |
Chen S, Zhao L. Association between the IL10 -1082A and gt; G (rs1800896) single-nucleotide polymorphism and atopic dermatitis: A systematic review and meta-analysis. Dermatitis 2013;24:161-5.
[Google Scholar]
|
| 58. |
Rousset F, Garcia E, Defrance T, Péronne C, Vezzio N, Hsu DH, et al. Interleukin 10 is a potent growth and differentiation factor for activated human B lymphocytes. Proc Natl Acad Sci U S A 1992;89:1890-3.
[Google Scholar]
|
| 59. |
Ala Y, Pasha MK, Rao RN, Komaravalli PL, Jahan P. Association of IFN-γ IL-10 cytokine ratio with nonsegmental vitiligo pathogenesis. Autoimmune Dis 2015;423490.
[Google Scholar]
|
| 60. |
Grimes PE, Morris R, Avaniss-Aghajani E, Soriano T, Meraz M, Metzger A. Topical tacrolimus therapy for vitiligo: Therapeutic responses and skin messenger RNA expression of proinflammatory cytokines. J Am Acad Dermatol 2004;51:52-61.
[Google Scholar]
|
| 61. |
Taher ZA, Lauzon G, Maguiness S, Dytoc MT. Analysis of interleukin-10 levels in lesions of vitiligo following treatment with topical tacrolimus. Br J Dermatol 2009;161:654-9.
[Google Scholar]
|
| 62. |
Sinha S, Kaler LJ, Proctor TM, Teuscher C, Vandenbark AA, Offner H, et al. IL-13-mediated gender difference in susceptibility to autoimmune encephalomyelitis. J Immunol 2008;180:2679-85.
[Google Scholar]
|
| 63. |
Eder L, Chandran V, Pellett F, Pollock R, Shanmugarajah S, Rosen CF, et al. IL13 gene polymorphism is a marker for psoriatic arthritis among psoriasis patients. Ann Rheum Dis 2011;70:1594-8.
[Google Scholar]
|
| 64. |
Chong KK, Chiang SW, Wong GW, Tam PO, Ng TK, Hu YJ, et al. Association of CTLA-4 and IL-13 gene polymorphisms with graves' disease and ophthalmopathy in chinese children. Invest Ophthalmol Vis Sci 2008;49:2409-15.
[Google Scholar]
|
| 65. |
Ying X, Zhang R, Yu S, Wu J, Wang H. Association of interleukin-13 SNP rs1800925 with allergic rhinitis risk: A meta-analysis based on 1,411 cases and 3169 controls. Gene 2012;506:179-83.
[Google Scholar]
|
| 66. |
Duffin KC, Freeny IC, Schrodi SJ, Wong B, Feng BJ, Soltani-Arabshahi R, et al. Association between IL13 polymorphisms and psoriatic arthritis is modified by smoking. J Invest Dermatol 2009;129:2777-83.
[Google Scholar]
|
| 67. |
Liao N, Zhao H, Chen ML, Xie ZF. Association of the IL-13 polymorphisms rs1800925 and rs20541 with chronic obstructive pulmonary disease risk: An updated meta-analysis. Medicine (Baltimore) 2017;96:e8556.
[Google Scholar]
|
| 68. |
Pertovaara M, Antonen J, Hurme M. Th2 cytokine genotypes are associated with a milder form of primary Sjogren's syndrome. Ann Rheum Dis 2006;65:666-70.
[Google Scholar]
|
| 69. |
Seyfizadeh N, Seyfizadeh N, Gharibi T, Babaloo Z. Interleukin-13 as an important cytokine: A review on its roles in some human diseases. Acta Microbiol Immunol Hung 2015;62:341-78.
[Google Scholar]
|
| 70. |
von Willebrand M, Köhler K, Alanko T, Laiho M, Saksela O. FGF-2 blocks TGF-beta1-mediated suppression of bcl-2 in normal melanocytes. Exp Dermatol 2005;14:202-8.
[Google Scholar]
|
| 71. |
Broen JC, Dieude P, Vonk MC, Beretta L, Carmona FD, Herrick A, et al. Polymorphisms in the interleukin 4, interleukin 13, and corresponding receptor genes are not associated with systemic sclerosis and do not influence gene expression. J Rheumatol 2012;39:112-8.
[Google Scholar]
|
| 72. |
Hancock A, Armstrong L, Gama R, Millar A. Production of interleukin 13 by alveolar macrophages from normal and fibrotic lung. Am J Respir Cell Mol Biol 1998;18:60-5.
[Google Scholar]
|
Fulltext Views
4,905
PDF downloads
2,739





