Translate this page into:
Molecular diagnostics in genodermatoses - simplified
2 Director-Research, SHARE, Andhra Prasesh, India
Correspondence Address:
Ravi N Hiremagalore
Department of Dermatology, Mediciti Institute of Medical Sciences, Ghanpur, Medchal Mandal, R. R. District - 501 401, Andhra Pradesh
India
| How to cite this article: Hiremagalore RN, Nizamabad N, Kamasamudram V. Molecular diagnostics in genodermatoses - simplified. Indian J Dermatol Venereol Leprol 2008;74:8-14 |
Abstract
The field of genetics in dermatology has progressed at an astonishing rate. Most of the known single gene disorders have at least been mapped to a particular chromosomal region and the causative genes have been identified and studied in many of them. However, most research work in genetics relating to genodermatoses has been confined to the western population. Very few reports, if any, have been published from Indian studies. A first step may be to develop a registry to link most of these cases providing a full description of the clinical phenotype. We would next need to attempt genetic analysis of these conditions thereby detecting any novel mutations in known and unknown genes different from the western population. This would help in designing indigenous assays appropriate to the Indian population. The review describes various techniques used in a molecular biology/ genetics laboratory with special focus on polymerase chain reaction (PCR), gene sequencing, genotyping and DNA micro arrays. Gene identification strategies have also been described with appropriate examples in dermatology. |
| Figure 4: Consider markers A and B 300kb apart linked to a particular disease phenotype. They are in linkage disequilibrium if they are always inherited together. However, by using more markers, C and D between them and studying more families, we can determine if there is any recombination between A and B. If there is recombination, then we analyze to determine which one is associated with the disease say for example A. Next we identify the next closest marker to B i.e. D and repeat the process to see if markers D and A are in linkage disequilibrium and the process is repeated till the smallest interval is found fl anked by markers in tight linkage disequilibrium |
 |
| Figure 4: Consider markers A and B 300kb apart linked to a particular disease phenotype. They are in linkage disequilibrium if they are always inherited together. However, by using more markers, C and D between them and studying more families, we can determine if there is any recombination between A and B. If there is recombination, then we analyze to determine which one is associated with the disease say for example A. Next we identify the next closest marker to B i.e. D and repeat the process to see if markers D and A are in linkage disequilibrium and the process is repeated till the smallest interval is found fl anked by markers in tight linkage disequilibrium |
 |
| Figure 3: Snap shot reaction This figure shows the basic steps in a Snapshot assay used in fluorescent genotyping. The DNA in the first step is denatured to separate the two strands. Next, the DNA is treated with unlabelled probe which stops one base short of the target polymorphism (SNP), which is G in this example. Then, the primer/probe is extended by one base using labeled dideoxynucleotides (ddNTPs), C in this case. This can be detected using an automated sequencer and appropriate software. Each ddNTP is detected separately as they are tagged differently with fluorescent dyes |
 |
| Figure 3: Snap shot reaction This figure shows the basic steps in a Snapshot assay used in fluorescent genotyping. The DNA in the first step is denatured to separate the two strands. Next, the DNA is treated with unlabelled probe which stops one base short of the target polymorphism (SNP), which is G in this example. Then, the primer/probe is extended by one base using labeled dideoxynucleotides (ddNTPs), C in this case. This can be detected using an automated sequencer and appropriate software. Each ddNTP is detected separately as they are tagged differently with fluorescent dyes |
 |
| Figure 2: Polymerase chain reaction In the first step, the two DNA strands (red and black) are separated. Next, the strands are treated with primer pairs (blue) which bind to the strands by complementary base pair binding in either direction. Then, with DNA polymerase, the primers are extended to form two new strands of DNA |
 |
| Figure 2: Polymerase chain reaction In the first step, the two DNA strands (red and black) are separated. Next, the strands are treated with primer pairs (blue) which bind to the strands by complementary base pair binding in either direction. Then, with DNA polymerase, the primers are extended to form two new strands of DNA |
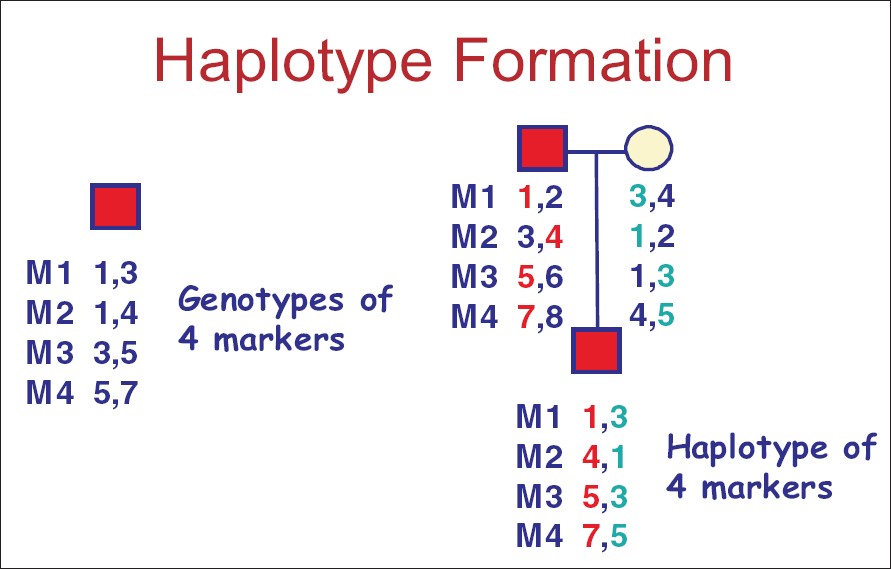 |
| Figure 1: Haplotype formation for markers M1 to M4 Numbers 1 to 7 are the different alleles for markers M1 to M4. Haplotype in the father include 1,3,5,7 and 2,4,6,8 whereas in the mother, 3,1,1,4 and 4,2,3,5 form the two haplotypes on both chromosomes. In the child, the haplotypes on the chromosomes include 1,4,5,7 and 3,1,3,5. By genotyping various markers in the parents and the child, we can identify the inheritance patterns of the haplotypes. In this example, haplotype 1,4,5,7 was inherited from the paternal chromosome whereas 3,1,3,5 from the mother. This kind of haplotype analysis has been extensively used in the genetic analyses of single gene diseases and complex traits with multifactorial etiology |
 |
| Figure 1: Haplotype formation for markers M1 to M4 Numbers 1 to 7 are the different alleles for markers M1 to M4. Haplotype in the father include 1,3,5,7 and 2,4,6,8 whereas in the mother, 3,1,1,4 and 4,2,3,5 form the two haplotypes on both chromosomes. In the child, the haplotypes on the chromosomes include 1,4,5,7 and 3,1,3,5. By genotyping various markers in the parents and the child, we can identify the inheritance patterns of the haplotypes. In this example, haplotype 1,4,5,7 was inherited from the paternal chromosome whereas 3,1,3,5 from the mother. This kind of haplotype analysis has been extensively used in the genetic analyses of single gene diseases and complex traits with multifactorial etiology |
Introduction
The field of genetics in dermatology has progressed at an astonishing rate. Most of the known single gene disorders have at least been mapped to a particular chromosomal region and the causative genes have been identified and studied in many of them. However, most research in genetics related to genodermatoses has been confined to the western population. Very few reports, if any, have been published from Indian studies. There are ever-increasing reports of novel mutations in known genes causing the same clinical syndrome in other ethnic populations. The review describes the various techniques used in a molecular biology/genetics laboratory with special focus on polymerase chain reaction (PCR), gene sequencing, genotyping and DNA micro arrays. Later, the various gene identification strategies are elaborated and finally, the methods of genetic testing have been discussed. Appropriate examples related to genodermatoses have been given to better explain the concepts throughout the review.
Need for Genetic Dissection of Common and Rare Genodermatoses
There is no doubt about the importance of genetic analyses of common and rare genodermatoses, the impact of which has been considerable in the west. The classical example is epidermolysis bullosa which has different clinical manifestations caused by mutations in a different set of genes. Specifically, it is now possible to make an accurate diagnosis of the type of epidermolysis bullosa. [1] Genotype-phenotype correlation is now possible, which helps determine prognosis in affected individuals. Such information is relevant to the medical care of the patient and also helps in evidence-based counseling of parents of patients to enable them to make informed decisions. Knowledge of mutations also makes DNA-based prenatal diagnosis feasible in families at risk of recurrence of an affected child in subsequent pregnancies [2] and helps us to detect the risk of transmission to offspring. Additionally, characterization of gene mutations has led to a major research initiative to develop somatic gene therapy with an emphasis on gene replacement. Studies are in progress for gene replacement therapy for recessive subtypes of epidermolysis bullosa (EB) associated with loss-of-function mutation on both alleles and an absence of the corresponding functional protein. [3]
What is its Relevance to India ?
A few of these genodermatoses have been published as clinical case reports while most of them go unattended. Moreover, Indians are ethnically quite different from Caucasians. A first step may be to develop a registry to link most of these cases providing a full description of the clinical phenotype. If this path were to be followed, we would need to attempt genetic analysis of these conditions thereby detecting any novel mutations in known and unknown genes different from the western population. This would help in designing indigenous assays appropriate to the Indian population and also help in understanding the genotype-phenotype differences from the rest of the world. In the long run, this would have great impact on gene therapy.
Terms Used in Genetics
- Locus : Location of a particular sequence of DNA (a gene or any fragment of DNA) on a chromosome.
- Allele : Alternative forms of a particular DNA sequence or a gene.
- Polymorphism : A locus exhibits at least two alleles with a frequency for one of them being greater than one per cent.
- Genotype : Alleles present at a specific locus.
- Phenotype : Manifestation of a particular genotype and the environment in which it is expressed.
- Allelic heterogeneity : Mutations in the same gene cause different clinical entities. As an example, mutations in the GJB3 gene cause erythrokeratodermia variabilis, autosomal dominant (AD) nonsyndromic sensorineural deafness and AD deafness with peripheral neuropathy. [4],[5],[6]
- Locus heterogeneity : Mutations in different genes produce the same phenotype. For example, epidermolysis bullosa simplex (EBS)-Koebner type can result from mutations in the keratin 5 and 14 gene. [7],[8]
-
Markers : Markers are segments of DNA that are different in different inherited chromosomes. They may be short terminal repeats (STRs) or single nucleotide polymorphisms (SNPs).
STR: CATGCACACACAACTG (Maternal)
CATGCACACAACTG (Paternal)
SNP: CATGGCTAGCTGA (Paternal)
CATGCCTAGCTGA (Maternal)
- Haplotype : A combination of alleles for several closely placed markers [Figure - 1].
Techniques in Molecular Biology
Polymerase Chain Reaction (PCR) [9],[10]
This is a technique of amplifying a specific segment of DNA. This is one of the basic techniques in any genetics or molecular biology laboratory.
Every PCR cycle is essentially composed of three steps [Figure - 2]:
a. Denaturation: In this step, the double-stranded DNA is separated into two single strands at specific temperatures greater than 90°C.
b. Annealing: In this step, the forward and reverse primers added to the reaction bind in opposite directions to the parent DNA with complementary base pair binding. Primers are oligonucleotides of 25-30 base length and complementary to the parent DNA template. Annealing occurs between 50-65°C.
c. Primer extension: DNA polymerase catalyzes the reaction where new bases are added to the primer, complementary to the DNA template. The reaction proceeds in both directions depending on the primer and occurs at an optimal temperature of 72°C.
The entire PCR process is done in a thermal cycler designed to provide optimal temperature conditions. Each cycle results in two new DNA segments which act as templates for the next cycle. Thus, a run of 35 cycles results in millions of copies of the same segment of DNA, which can be detected on an agarose gel. PCR can be done using either whole blood or skin biopsy specimens. It is a very sensitive technique and has immense applications in clinical medicine and research. PCR is widely being used for the diagnosis of pathogens when extremely low quantities are present in tissues. It is one of the first steps in identifying polymorphisms in various genodermatoses and has wide applications in genetic analysis of complex traits like psoriasis. A limitation of this technique is the potential for laboratory contamination of the DNA sample by trace amounts of the PCR product, which can give rise to misleading results particularly when used to detect microbial agents. There are two modifications to PCR; one is called RT-PCR (reverse transcriptase-PCR), which uses RNA instead of DNA as the starting point while the other is called Real-Time PCR, which is a quantitative method. This method is more popular for detecting microbial agents and for gene expression studies.
This is a technique of identifying the exact sequence of nucleotides in a specified DNA segment using radiolabeled bases or in an automated fluorescence-based sequencer. DNA sequencing involves the following steps:
a. An initial PCR
b. After amplification, the PCR product is treated with DNA polymerase, ddNTPs (dideoxy nucleoside triphosphates, terminator nucleotides), each of the four types tagged with different fluorescent dyes and a primer. As the reaction proceeds, the primer binds to the DNA to be sequenced and DNA polymerase synthesizes a new complementary strand. The strand is randomly terminated by the incorporation of the labeled ddNTPs tagged with different fluorescent dyes, thus enabling the identification of the segment. These fragments are identified using capillary tubes in an automated sequencer and read using appropriate software programs. Direct sequencing has been used to screen polymorphisms in a gene which may be pathogenic or a normal variation in the population. It forms the basis for research into the genetics of complex diseases like psoriasis. Recently, shotgun sequencing has led to the identification that HLA-Cw6 is the PSORS1 gene. [11] Sequencing requires high-quality DNA and certain areas of the genome particularly rich in G-C areas are difficult to sequence.
Genotyping
This technique involves determining whether a polymorphism exists in a heterozygous or homozygous form in an individual, thereby determining the allele frequency in the given population. Genotype-phenotype correlation can help determine the risk of developing the disease and also its prognosis. Various methods of genotyping include snapshot assays and real-time assays. Snapshot assays detect single nucleotide polymorphisms (SNPs) using fluorescent dyes. [12] The initial step is a PCR. The PCR product is treated with appropriately designed probes, which are basically oligonucleotides falling one base short of the target SNP. The probes bind to the product and they are extended by one base at optimal temperature conditions. Depending on the target SNP, the appropriate fluorescently tagged base is added and detected using an automated sequencer [Figure - 3].
Southern Blotting : [10] Genomic DNA extracted from cells can be cut with a suitable restriction enzyme and then fractionated according to size through an agarose gel in an electric field. The DNA is then transferred onto a nitrocellulose membrane and then hybridized with a radiolabeled probe complementary to the target sequence. The membrane is then washed to remove excess probe and placed next to a radiosensitive film overnight. The difference in the bands seen on the autoradiograph reflects the size of the DNA fragment containing the gene of interest. A similar procedure done using RNA to study expression profiles is called Northern blotting.
DNA micro arrays : DNA microarrays are the latest in a line of molecular biology techniques that exploit the unique feature of single-stranded DNA to hybridize to complementary DNA sequences, thereby permitting sequence-specific identification of DNA. This is superior to conventional hybridization techniques in that thousands of genes can be studied on one platform.
Each microarray experiment consists of five discrete steps: [13],[14],[15],[16],[17],[18],[19]
- Fabrication of the DNA microarray, which begins with the application of probes in an arrayed manner on either nylon membrane or glass. The probes are either fluorescent dye-labeled or P 32 -labeled. The biological sample, which is usually RNA, is extracted from a tissue. Typically, 10-40 µg of high-quality RNA is required.
- Hybridization of labeled nucleic acid with array.
- Signal detection using confocal microscopy if fluorescent dye-labeled probes are used or with phosphorimager screen if P 32 -labeled probes are used.
- Data processing and analysis using appropriate software.
Applications in Dermatology
- The main application is the molecular classification and genome-based diagnosis of diseases, mostly of neoplasms [20] and also inflammatory dermatoses via gene expression profiling. Diseases like scleroderma, alopecia areata and psoriasis have been extensively studied using this technique. [21],[22],[23]
- Effects of ultraviolet (UV) light on the skin have also been studied. [24]
- Mutational analysis of genodermatoses and genotyping and identifying polymorphisms.
Oligonucleotide arrays have been used for this purpose. [25] These nucleotides comprise sequences of all known genetic mutations for a particular disease. Such array-based detection is available for cystic fibrosis with 37 known mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene as well as for hereditary breast and ovarian cancer. [25] A comparable assay could replace the labor-intensive and often ambiguous electron microscopic diagnosis of the epidermolysis bullosa group. This technology requires the knowledge of known mutations, which is presently available only for the western population. Similar such studies involving the Indian population would aid in the design of our own indigenous array-based assays.
Gene Identification Strategies [26]
Advances in the application of molecular biology to the genetics of human disease have largely been confined to disorders characterized by Mendelian inheritance pattern. There are two strategies to identify disease-causing genes:
- Functional cloning : It is based on the fact that there is a known biochemical defect causing the disorder. A candidate gene is thus identified based on prior knowledge of the pathophysiology and the gene product is characterized. The candidate gene is screened for mutations and studied in the affected patients. This approach is exemplified in the identification of mutations in the transglutaminase 1 (TGM1) gene as the cause of lamellar ichthyosis. [27],[28] This enzyme is involved in the formation of the cornified envelope and patients with lamellar ichthyosis have undetectable TGM1 activity in frozen skin specimens. [29] In another instance, when the tyrosinase and tyrosinase-related genes were identified; they were obvious candidates to examine for mutations in individuals with albinism. [30]
-
Positional cloning : This process identifies the gene based only on its chromosomal location. No prior information about the gene and its product are known or required. Steps involved in positional cloning are as follows:
a. The first step is chromosomal assignment of the region containing the defective gene by a method called linkage analysis. The aim is to identify the chromosomal region that is transmitted with the disease phenotype, subsequently linking the disease to the genetic locus which harbors the gene. Genome-wide scans using several markers are analyzed by PCR for this purpose. Linkage analysis requires a proper definition of the disease phenotype, pedigrees with enough power (a single large or several small families is good enough), a dense map of polymorphic markers (either SNPs or microsatellites) placed at 10-cm intervals ( i.e ., about 300 markers) and application of statistical methods.
b. Analyses of data from genome-wide scans are done using the logarithmic odds (LOD) score to determine linkage. An LOD score ≥3 is suggestive of definitive linkage while a score of < 2 rules it out.
c. The next step is refinement of the linkage interval. This can be done by finding loss of heterozygosity or a chromosomal abnormality in a subset of individuals. In most cases however, this can be achieved by increasing the sample size and the number of markers analyzed within the interval. Formation and analysis of haplotypes is essential for this step [Figure - 1]. The goal is to identify the boundaries of the smallest interval, defined by closely placed markers that show recombination with the disease phenotype. The rationale is that the closer together the two markers are, the less frequently they will be separated by recombination. They are thus said to be in linkage disequilibrium. In the [Figure - 4], consider markers A and B 300 kilobases (kb) apart linked to a particular disease phenotype. They are in linkage disequilibrium if they are always inherited together. However, by using more markers between them and studying more families, we can determine if there is any recombination between A and B. If there is recombination, then we determine which one is associated with the disease, say for example, A. Next we identify the next closest marker to B and repeat the process to see if the new marker and marker A are in linkage disequilibrium and the process is repeated till the smallest interval is found flanked by markers in tight linkage disequilibrium.
d. After refinement of the linkage interval, identification of the actual gene begins. In the positional candidate gene approach, the gene is mapped to the locus interval using information from the human genome project. For example, in Darier′s disease, the locus was localized to chromosome 12 by linkage. [31],[32] After refinement, several genes were identified among which ATP2A2 encoding the calcium pump SERCA2 was considered a good candidate due to the role of Ca 2+ in epithelial junctions and cell differentiation. In another study, linkage to chromosome 3 was established for Hailey-Hailey disease. [33] In this interval, only one gene ATPA2C1 showed homology to calcium ATPases and was subsequently found mutated in Hailey-Hailey patients. [34],[35]
e. If no candidate gene is found, positional cloning is done to identify new genes in the interval.
f. Finally, after the gene is identified, it is sequenced to identify polymorphisms. These are tested for association with the disease by comparing affected and unaffected members in the patient′s family.
Genetic Testing in Individuals [36]
How much information a genetic test can give depends on the state of knowledge about the gene involved, but in principle, laboratory genetic testing can be done in two ways:
- Gene tracking : This was the first type of DNA diagnostic method used. A group of linked markers are used in a family to determine whether or not the affected individual inherited the high-risk chromosome from a heterozygous parent. In this method, it is not the genotype of the marker that is important but its segregation pattern in the family.
- Direct mutation testing : It is almost always a PCR-based technique. The tissue samples commonly used are whole blood, buccal scrapes, chorionic villi and even skin biopsy specimens. RNA-based RT-PCR has distinct advantages over the DNA-based PCR but is less convenient to obtain and work with. This method is useful in diseases with limited allelic heterogeneity where the same mutation causes the disease in all patients. Some of the commonly used techniques include testing for the presence or absence of restriction sites, allele-specific oligonucleotide hybridization, PCR using allele-specific primers (amplification refractory mutation system [ARMS] test). In diseases with extensive allelic heterogeneity, mutation scanning is done first to identify the mutation in the family. Once one affected member is tested, then the same mutation can be tested in other affected members of the same family. For research, direct sequencing and heteroduplex/single-stranded conformational polymorphism (SSCP) are most commonly used whereas denaturing gradient gel electrophoresis (DGGE) is used for diagnostic purposes. Other techniques include southern blotting, protein truncation test (PTT) and more recently, oligonucleotide arrays. All of these methods have the limitation of being laborious and expensive. They detect differences between the patient′s sequence and the published normal sequence but do not distinguish between pathogenic and chance nonpathogenic changes. Homozygosity mapping has been reported recently as a cost-effective screening tool to direct subsequent molecular diagnosis of genetically heterogenous genodermatosis. This is particularly relevant in consanguineous populations as seen in some parts of India. The two commonly studied diseases are junctional epidermolysis bullosa and congenital recessive ichthyosis.
Conclusion
Gene identification strategies have solved the genetic mysteries underlying several common genodermatoses. However, newer and rarer conditions need to be elucidated at the molecular level. It seems likely that eventually oligonucleotide arrays will replace most other methods for routine mutation scanning of the more common diseases and automated sequencing will be increasingly used for rarer diseases. In India, we are yet to crack the genetic puzzle of most genodermatoses. Moreover, our population being genetically diverse, mandates the establishment of an indigenous mutation database of these conditions before assays can be designed for diagnostic work.
| 1. |
Uitto J, Pulkinin L. Molecular genetics of heritable blistering disorders. Arch Dermatol 2001;137:1458-61.
[Google Scholar]
|
| 2. |
Pfendner EG, Nakano A, Pulkkinen L, Christiano AM, Uitto J. Prenatal diagnosis for Epidermolysis bullosa: A study of 144 consecutive pregnancies at risk. Prenat Diagn 2003;23:447-56.
[Google Scholar]
|
| 3. |
Khavari PA, Rollman O, Vahlquist A. Cutaneous gene transfer for skin and systemic diseases. J Intern Med 2002;252:1-10.
[Google Scholar]
|
| 4. |
Richard G, Smith LE, Bailey RA, Itin P, Hohl D, Epstein EH Jr, et al. Mutations in the human connexin gene GJB3 cause erythrokeratodermia variabilis. Nat Genet 1998;20:366-9.
[Google Scholar]
|
| 5. |
Xia JH, Liu CY, Tang BS, Pan Q, Huang L, Dai HP, et al. Mutations in the gene encoding gap junction protein beta-3 associated with autosomal dominant hearing impairment. Nat Genet 1998;20:370-3.
[Google Scholar]
|
| 6. |
Lopez-Bigas N, Olive M, Rabionet R, Ben-David O, Martνnez-Matos JA, Bravo O, et al. Connexin 31(GJB3) is expressed in the peripheral and auditory nerves and causes neuropathy and hearing impairment. Hum Mol Genet 2001;10:947-52.
[Google Scholar]
|
| 7. |
Dong W, Ryynanen M, Uitto J. Identification of a leucine to proline mutation in the keratin 5 gene in a family with generalized koebner type of EBS. Hum Mutat 1993;2:94-102.
[Google Scholar]
|
| 8. |
Bonifas JM, Rothman AL, Epstein EH Jr. Epidermolysis bullosa simplex: Evidence in two families for keratin gene abnormalities. Science 1991;254:1202-5.
[Google Scholar]
|
| 9. |
Vogel J, Yee C, Darling T. Molecular biology. In : Bolognia JL, Jorizzo JL, Rapini RP, editors. Dermatology, 1 st ed. Mosby: New York; 2003. p. 49-63.
[Google Scholar]
|
| 10. |
Rees JL. Molecular biology. In : Burns T, Breathnach S, Cox N, Griffiths C, editors. Rook's Textbook of Dermatology: Molecular Biology. Blackwell Science: Oxford; 2004. p. 8.1-8.24.
[Google Scholar]
|
| 11. |
Nair RP, Stuart P, Nistor I, Hiremagalore R, Chia NV, Jenisch S, et al. Sequence and Haplotype Analysis supports HLA-C as the Psoriasis susceptibility 1 gene. Am J Hum Genet 2006;78:827-51.
[Google Scholar]
|
| 12. |
Schena M, Heller RA, Theriault TP, Konrad K, Lachenmeier E, Davis RW. Microarrays: Biotechnology's discovery platform for functional genomics. Trends Biotechnol 1998;16:301-6.
[Google Scholar]
|
| 13. |
Bowtell DD. Options available-from-start to finish-for obtaining expression data by micro array. Nat Genet 1999;21:25-32.
[Google Scholar]
|
| 14. |
Brown PO, Botstein D. Exploring the new world of the genome with DNA micro arrays. Nat Genet 1999;21:33-7.
[Google Scholar]
|
| 15. |
Khan J, Bittner ML, Chen Y, Meltzer PS, Trent JM. DNA micro array technology: The anticipated impact on the study of human disease. Biochem Biophys Acta 1999;1423:M17-28.
[Google Scholar]
|
| 16. |
Duggan DJ, Bittner M, Chen Y, Meltzer P, Trent JM. Expression profiling using cDNA micro arrays. Nat Genet 1999;21:10-4.
[Google Scholar]
|
| 17. |
Maughan NJ, Lewis FA, Smith V. An introduction to arrays. J Pathol 2001;195:3-6.
[Google Scholar]
|
| 18. |
Wessagowit V, South AP. Dermatological applications of DNA micro array technology. Clin Exp Dermatol 2002;27:485-92.
[Google Scholar]
|
| 19. |
Khan J, Wei JS, Ringner M, Saal LH, Ladanyi M, Westermann F, et al . Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat Med 2001;7:673-9.
[Google Scholar]
|
| 20. |
Zhou X, Tan FK, Xiong M, Milewicz DM, Feghali CA, Fritzler MJ, et al . Systemic sclerosis (scleroderma): Specific autoantigen genes are selectively overexpressed in scleroderma fibroblasts. J Immunol 2001;167:7126-33.
[Google Scholar]
|
| 21. |
Carroll JM, McElwee KJ, King LE Jr, Bryne MC, Sundberg JP. Gene array profiling and immunomodulation studies define a cell-mediated immune response underlying the pathogenesis of alopecia areata in a mouse model and humans. J Invest Dermatol 2002;119:392-402.
[Google Scholar]
|
| 22. |
Nomura I, Gao B, Boguniewicz M, Darst MA, Travers JB, Leung DY. Distinct patterns of gene expression in the skin lesions of atopic dermatitis and psoriasis: A gene micro array analysis. J Allergy Clin Immunol 2003;112:1195-202.
[Google Scholar]
|
| 23. |
Li D, Turi TG, Schuck A, Freedberg IM, Khitrov G, Blumenberg M. Rays and micro arrays: The transcriptional program in the response of human epidermal keratinocytes to UVB illumination. FASEB J 2001;15:2533-5.
[Google Scholar]
|
| 24. |
Hacia JG. Resequencing and mutational analysis using oligonucleotide micro arrays. Nat Genet 1999;21:42-7.
[Google Scholar]
|
| 25. |
Strachan T, Read AP. Identifying human disease genes. In : Human Molecular Genetics. Bios Scientific Publishers: Oxford; 1996. p. 351-75.
[Google Scholar]
|
| 26. |
Huber M, Rettler I, Bernascomi K, Frenk E, Lavrijsen SP, Ponec M, et al . Mutations of keratinocyte transglutaminase in Lamellar ichthyosis. Science 1995;267:525-8.
[Google Scholar]
|
| 27. |
Rusell LJ, Digiovanna JJ, Rogers GR, Steinert PM, Hashem N, Compton JG, et al . Mutations in the gene for transglutaminase 1 in autosomal recessive lamellar ichthyosis. Nat Genet 1995;9:279-83.
[Google Scholar]
|
| 28. |
Hold D, Aeschlimann D, Huber M. In vitro and rapid in situ transglutaminase assays for congenital ichthyosis- a comparative study. J Invest Dermatol 1998;110:268-71.
[Google Scholar]
|
| 29. |
Spritz RA. Molecular genetics of oculocutaneous albinism. Hum Mol Genet 1994;3:1469-75.
[Google Scholar]
|
| 30. |
Craddock N, Dawson E, Burge S, Parfitt L, Mant B, Roberts Q, et al . The gene for Darier's disease maps to chromosome 12q23-q24.1. Hum Mol Genet 1993;2:1941-3.
[Google Scholar]
|
| 31. |
Bashir R, Munro CS, Mason S, Stephenson A, Rees JL, Strachan T. Localization of a gene for Darier's disease. Hum Mol Genet 1993;2:1937-9.
[Google Scholar]
|
| 32. |
Ikeda S, Welsh EA, Peluso AM, Leyden W, Duvic M, Woodley DT, et al . Localization of the gene whose mutations underlie hailey-Hailey disease to chromosome 3q. Hum Mol genet 1994;3:1147-50.
[Google Scholar]
|
| 33. |
Hu Z, Bonifas JM, Beech J, Bench G, Shigihara T, Ogawa H, et al . Mutations in ATPA2C1, encoding a calcium pump, cause Hailey-Hailey disease. Nat Genet 2000;24:61-5.
[Google Scholar]
|
| 34. |
Sudbrak R, Brown J, Dobson-Stone C, Carter S, Ramser J, White J, et al . Hailey-Hailey disease is caused by mutations in ATPA2C1 encoding a novel Calcium pump. Hum Mol Genet 2000;9:1131-40.
[Google Scholar]
|
| 35. |
Strachan T, Read AP. Genetic testing in individuals and populations. In : Human Molecular Genetics. Bios Scientific Publishers: Oxford; 1996. p. 401-25.
[Google Scholar]
|
| 36. |
Mizrachi-Koren M, Shemer S, Morgan M, Indelman M, Khamaysi Z, Petronius D, et al . Homozygosity mapping as a screening tool for the molecular diagnosis of hereditary skin diseases in consanguineous populations. J Am Acad Dermatol 2006;55:393-401.
[Google Scholar]
|
Fulltext Views
5,346
PDF downloads
4,754





